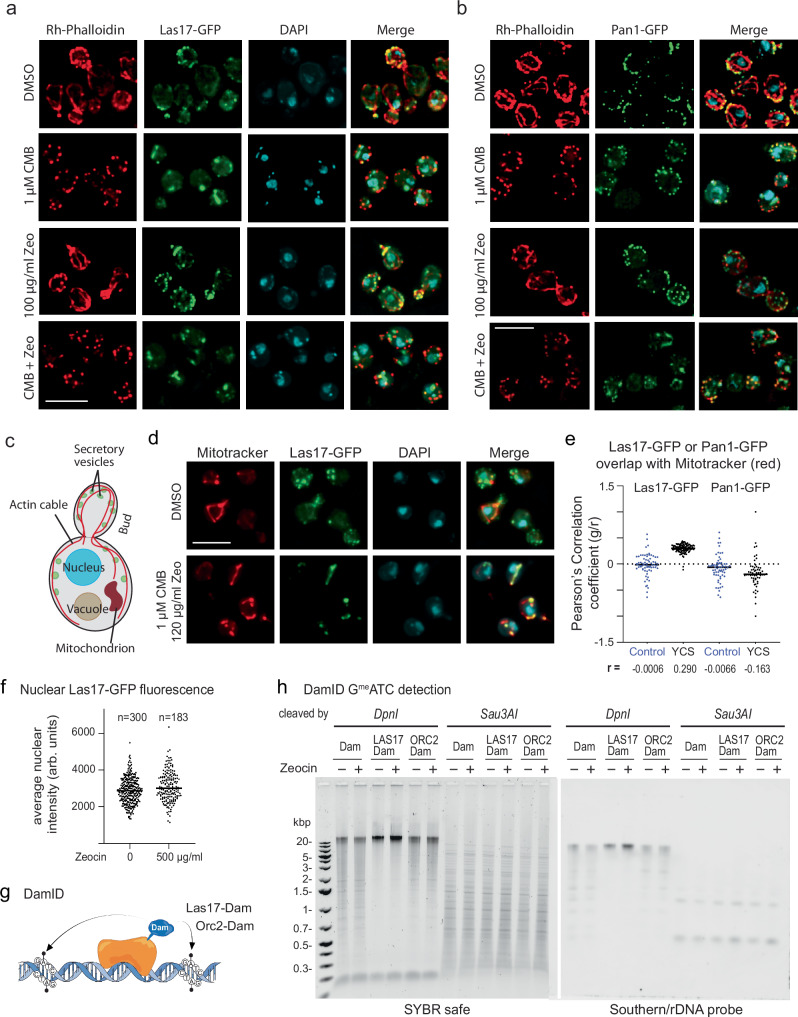
Fig. 5. Pan1 and Las17 do not shift to the nucleus in the presence of Zeocin or CMB + Zeocin.
a Yeast expressing Las17-GFP (GA-6804) was treated 1.5 h with CMB (1 μM), with Zeocin (100 μg/ml) or both. After fixation, DAPI (blue) and F-actin (Rh-phalloidin, red) were captured by spinning disk confocal microscopy. Images are maximum-intensity projections of focal stacks acquired in each channel (see Methods). Bar = 5 µm. b Yeast expressing Pan1-GFP (GA-6764) was treated and stained as in a. DAPI alone is not shown. Bar = 5 µm. c Scheme of a budding yeast cell illustrating structures of interest. d Las17-GFP expressing cells were treated as in a, but stained with 20 nM Mitotracker Red CMXRos (Thermo Scientific) and DAPI (blue). Colocalization of Las17-GFP and mitochondria is quantified in e. Bar = 5 µm. e Yeast expressing Las17-GFP or Pan1-GFP (see a, b) were split and treated 1.5 h either with DMSO (Control, blue) or 1 µM CMB and 150 µg/ml Zeocin (YCS, black). Colocalization of Las17-GFP or Pan1-GFP (green) and Mitotracker (red) was quantified by determining the Pearson correlation coefficient in each single plane of an image stack. n = 118 nuclei for Las17-YCS, all others n = 78; line = median. r values show a robust correlation of Las17 with mitochondria. Repeated twice. Quantitations in Source Data file. f Las17-GFP is not enriched in nuclei on Zeocin. Las17-GFP (green) expressing cells were treated ± 500 μg/ml Zeocin for 1 h at 30 °C, fixed and counterstained with DAPI (blue). Image stacks (green and UV) were analyzed for nuclei spanning at least 5 planes. Nuclear GFP intensity was measured per plane and averaged across ≥5 planes for mean intensity per nucleus, plotted as arbitrary GFP units. n = 300 nuclei without Zeo, mean and S.D. = 2861±700; n = 183 with Zeo, mean and S.D. = 3002±805. Unpaired two-tailed T test with Welch’s correction, p = 0.051. See Source Data Files. g Concept of DamID mapping71 of Las17- and Orc2-Dam fusions, derived from an existing sketch72. h Wild-type (GA-1981) cells expressing Dam, Las17-Dam, or Orc2-Dam were treated ±300 μg/ml Zeocin for 1 h at 30 °C. Genomic DNA digested with DpnI (cleaves at GmATC) or Sau3AI (cleaves GATC, methylation indifferent), was run on 1% agarose gels; stained with SYBR safe or rDNA by Southern blot (see Methods).