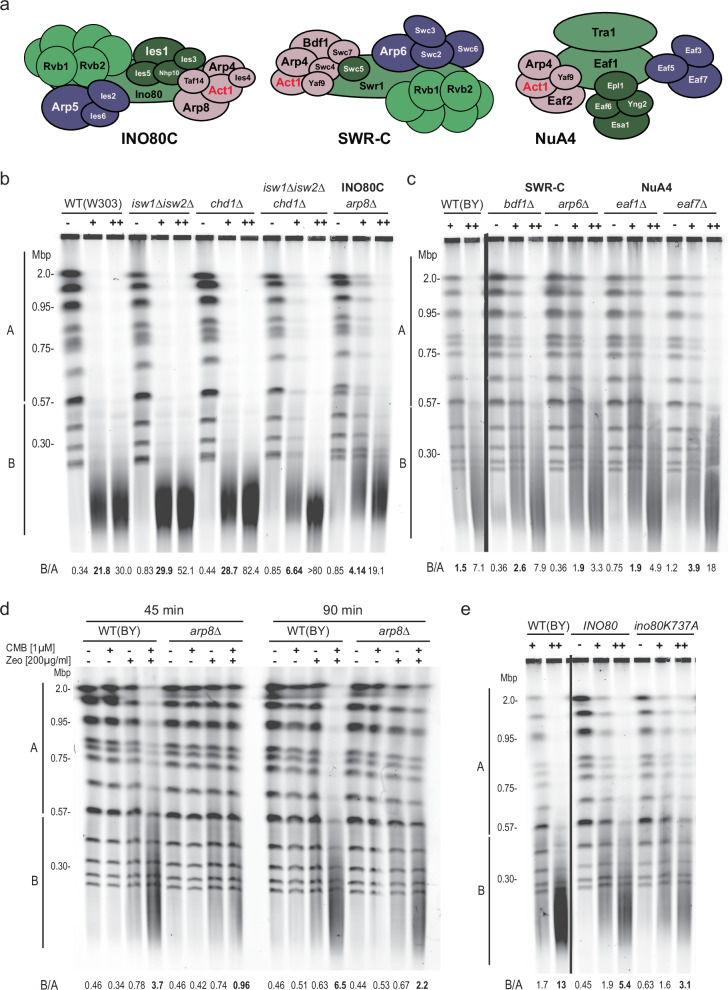
Fig. 8. Functional INO80C is required for efficient YCS.
a Sketches of subunit composition of the S. cerevisiae INO80C, SWR-C and NuA4 chromatin remodelers and modifiers each containing actin as a core subunit12–15. Subunit names are from budding yeast. b Individual ISWI and CHD remodelers are not needed for YCS, while loss of Arp8, a unique INO80C subunit confers partial resistance. Exponential cultures of wild-type (WT) W303 (GA-9875); isw1Δ,isw2Δ (GA-9878); chd1Δ (GA-9879); isw1Δ isw2Δ chd1Δ (GA-9882) and arp8Δ (GA-9848) were treated for 1.5 h with 0.5 μM CMB and 50 μg/ml Zeocin (+) or 1 μM CMB and 100 μg/ml Zeocin (++), prior to processing for CHEF gel analysis. Quantitation as in Fig. 1b. c Isogenic strains in the BY background lacking indicated remodeler subunits of SWR-C or NuA4 were tested in the YCS assay. Note that this background is more resistant to Zeocin than W303. BY4733 (GA-2263), bdf1Δ (GA-9953), arp6Δ (GA-2319), eaf1Δ (GA-9954), eaf7Δ (GA-9955) were grown to exponential phase and treated for 1.5 h with DMSO (−), 0.5 µM CMB and 80 µg/ml Zeocin (+) or 1 µM CMB and 125 µg/ml Zeocin (++), prior to CHEF gel analysis. Quantitation as in Fig. 1b. The eaf7Δ mutant showed slight resistance at low concentrations of Zeocin/CMB, while arp6Δ and eaf1Δ mutants showed minor resistance at the higher concentration. Except for INO80C, the minor effects were inconsistent for a given remodeler. d Deletion of arp8 delays YCS consistent with a requirement for functional INO80C for polymerase processivity during LP-BER. The indicated isogenic strains (ARP8 + = GA-9247; arp8Δ = GA-9250) were incubated with the indicated compounds for either 45 or 90 min, prior to processing for CHEF gel analysis. Quantitation is as in Fig. 1b. At 45 min ARP8+ started to show fragmentation, unlike arp8Δ. The experiment was repeated twice. e The ATPase mutant in the catalytic subunit ino80 (K737A) confers resistance to YCS. Performed in the BY background, ino80Δ (GA-2264) cells bearing Cen-plasmids expressing WT INO80 or ino80 K737A were treated for 1.5 h with 0.5 μM CMB and 80 μg/ml Zeocin (+) or 1 μM CMB and 125 μg/ml Zeocin (++); CHEF gel analysis and quantitation as Fig. 1b. Experiment repeated twice.