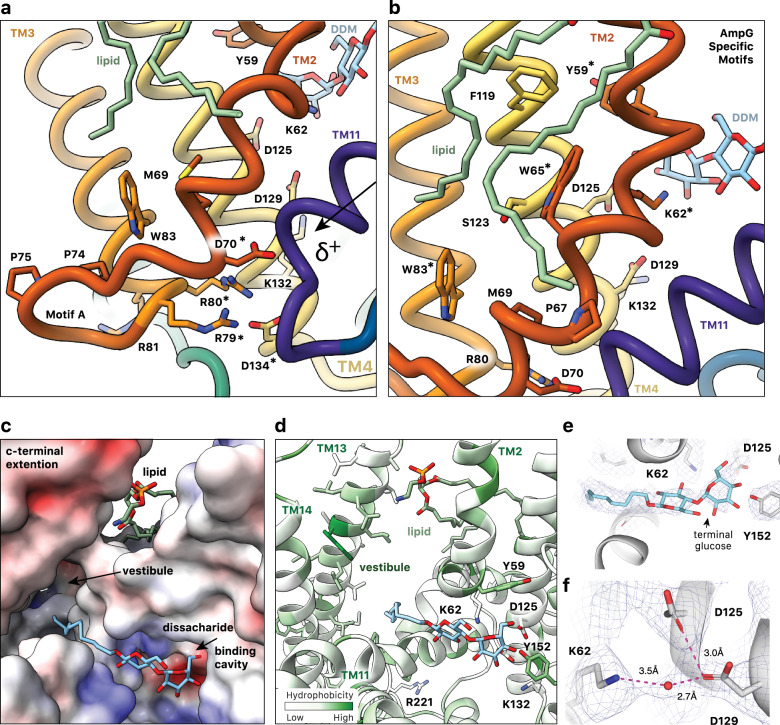
Fig. 4. Substrate binding cavity of outward facing E. coli AmpG.
a AmpG motif A stabilizes the outward conformation with the Asp70 carboxylate forming an electrostatic interaction with the N-terminal helix dipole of TM11, likely enhanced by coordination with Arg79, Arg80, and Asp134. Colored as in Fig. 1d, with mentioned residues marked by *. b Conserved Trp65 and Trp83 in the motif A adaption stabilize the kink in TM2, creating an expanded binding cavity and positioning key residues e.g., Tyr59 and Lys62 for substrate binding. The observed PE lipid extends across the aromatic and nonpolar residues on TM2 and TM4. c Electrostatic surface of the periplasmic substrate binding cavity of AmpG with bound DDM. The maltose sits near an electronegative pocket arising from conserved Asp125 and Asp134, while the hydrophobic acyl chain points into the hydrophobic vestibule between the MFS helical bundle and additional C-terminal helices TM13 and 14. d AmpG substrate binding cavity colored by hydrophobicity. Side chains of disaccharide binding residues shown as sticks and labeled. DDM and lipid are shown as in a. e Density in the binding cavity of AmpG of the modeled bound DDM. f Density consistent with a water coordinated between Lys62, Asp125, and Asp129.