Abstract
Mollusk-hunting (molluscivorous) cone snails belong to a monophyletic group in Conus, a genus of venomous marine snails. The molluscivorous lineage evolved from ancestral worm-hunting (vermivorous) snails ∼18 Ma. To enable the shift to a molluscivorous lifestyle, molluscivorous cone snails must solve biological problems encountered when hunting other gastropods, namely: (i) preventing prey escape and (ii) overcoming the formidable defense of the prey in the form of the molluscan shell, a problem unique to molluscivorous Conus. Here, we show that χ-conotoxins, peptides exclusively expressed in the venoms of molluscivorous Conus, provide solutions to the above problems. Injecting χ-conotoxins into the gastropod mollusk Aplysia californica results in impaired locomotion and uncoordinated hyperactivity. Impaired locomotion impedes escape, and a hyperactive snail will likely emerge from its shell, negating the protection the shell provides. Thus, χ-conotoxins are an evolutionary innovation that accompanied the emergence of molluscivory in Conus and provide solutions to problems posed by hunting other snails.
Keywords: conotoxins, mollusk-hunting cone snails, evolutionary innovation, emergence of complex phenotypes, χ-conotoxin
Introduction
The emergence of evolutionary novelties in a particular lineage may promote the diversification of its taxa. This macro-evolutionary change is accompanied by genetic and molecular innovations rarely described in detail due to the complex multigene dependence of many of these traits. Genotype-to-phenotype correlation in animal venoms is an ideal system for detecting genetic and molecular innovations important for lineage diversification. In this report, we demonstrate how a shift in prey preference by a biodiverse lineage of predators, the cone snails, was accompanied by genetic and molecular level adaptations.
There are ∼1,000 living species of cone snails, which are conventionally divided into three broad categories based on their prey: worm-hunting (vermivorous), fish-hunting (piscivorous), and mollusk-hunting (molluscivorous; Kohn 1959; Rockel et al. 1995; Terlau and Olivera 2004). All cone snails use venom to subdue prey. This venom is primarily composed of small, gene-encoded peptides called conopeptides or conotoxins that can be grouped into gene superfamilies (Terlau and Olivera 2004). Conotoxins from piscivorous snails have been the primary focus of research due to their therapeutic potential. Similarly, much research has been conducted to elucidate how certain classes of conotoxins enable these snails to hunt fish (Terlau et al. 1996; Olivera 1997; Olivera et al. 2015). In contrast, very little is known about the roles of conotoxins in molluscivorous and vermivorous species. For example, it is unclear which conotoxins address biological issues unique to molluscivory, such as the defensive shell of their prey.
Before a predator can successfully feed on gastropods, the prey of all molluscivorous cone snails, the following challenges must be resolved: (i) preventing prey escape and (ii) overcoming their main defense, the shell. Other predatory lineages of mollusks (e.g. Muricidae and Naticidae) solve this problem by pinning down their prey, drilling a hole into the shell, and sucking out the soft tissues with their proboscis (Kowalewski 2004; Pahari et al. 2016). However, cone snails lack such mechanisms, and must instead rely on venom.
Molluscivorous Conus, like all cone snails, uses a complex venom to capture prey (Olivera et al. 1985; Olivera 2002, 2006). Most well-characterized cone snail venom components target ion channels (voltage-gated and ligand-gated) or receptors (G protein–coupled receptors and receptor tyrosine kinases; Olivera 1997; Craig et al. 1999; Terlau and Olivera 2004; Lewis et al. 2012; Safavi-Hemami et al. 2015; Espino et al. 2018). The venom of each species contains several hundred different bioactive components, encoded by genes that are subject to accelerated evolution (Duda and Palumbi 1999; Phuong and Mahardika 2018). As a consequence, every cone snail species has its distinctive complement of conotoxins that are present in venom, with minimal overlap between conotoxins found in the venom of any two different species (Barghi et al. 2015; Li et al. 2018). However, functionally homologous conotoxins that have diverged from one another at the amino acid level can be found between closely related species (Olivera et al. 2015).
The most intensively studied venom component in the repertoire of a mollusicvorous Conus is a peptide now widely known as χ-conotoxin MrIa (χ-MrIa) of the T-superfamily (Balaji et al. 2000; McIntosh et al. 2000; Sharpe et al. 2001). This conotoxin inhibits the human norepinephrine transporter (hNET) and was shown to have analgesic activity (McIntosh et al. 2000; Sharpe et al. 2001) and an analog of χ-MrIa reached Phase II human clinical trials (Nielsen et al. 2005).
In this work, we provide evidence that χ-conotoxins are expressed exclusively by molluscivorous Conus. We further investigate the rationale for such a unique expression pattern, focusing primarily on the role of these conotoxins in predation. We provide evidence that these conotoxins are important for prey capture, specifically for solving two biological problems: preventing prey escape and forcing the prey out of its shell. The evolution of the χ-conotoxins may have been one of the key innovations accompanying the shift in prey type from worms to mollusks, allowing molluscivorous Conus to successfully diversify into ∼100 extant species (WoRMSEditorialBoard 2023) and colonize the tropical waters of the Indo-Pacific.
Results
Molluscivory Evolved Once in the Evolutionary History of Cone Snails
Previous research has shown that each of the major Conus feeding groups falls into distinct phylogenetic lineages, of which most have retained the ancestral worm-hunting trait (Duda et al. 2001; Duda and Palumbi 2004). Several analyses have shown that piscivory evolved multiple times (Duda et al. 2001; Espiritu et al. 2001; Puillandre et al. 2014; Lee and Park 2022). In contrast, only one lineage of molluscivores has been identified using traditional phylogenetic markers (e.g. 12SrDNA, 16SrDNA, CO1; Duda et al. 2001; Espiritu et al. 2001; Puillandre et al. 2014). Recently, we have shown that housekeeping genes (HKGs) prove useful in resolving the phylogenetic relationships in Conoidea, a large superfamily of venomous gastropods that includes cone snails (Chase et al. 2022). Here, we used the same approach to reconstruct the cone snail phylogeny and to determine the number of lineages that gave rise to each of the feeding groups.
To ensure a robust phylogenetic framework, we employed different strategies for reconstructing the evolutionary history of Conus lineages: gene tree analyses (maximum likelihood approaches, with concatenated and partitioned by locus datasets in IQ-Tree2, and species tree analysis using ASTRAL-III), and divergence timing analyses with a Bayesian framework (BEAST2). A concatenated dataset provided a preliminary insight into the overall tree topology (see supplementary fig. S1, Supplementary Material online). We followed that analysis by conducting individual gene tree reconstructions for each of the 86 markers (i.e. partition by locus, see supplementary fig. S2, Supplementary Material online). However, given the rapid radiation and complex branching patterns within the Conus genus (Puillandre et al. 2014; Uribe et al. 2017; Abalde et al. 2019), particularly among closely related species (Abalde et al. 2019), addressing gene tree discordance was crucial. ASTRAL-III (Zhang et al. 2018) was thus utilized to generate a species tree that effectively accounts for gene tree discordance, providing a more accurate reflection of cone snail evolutionary relationships (supplementary fig. S3, Supplementary Material online).
The choice of the species tree as the primary topology for subsequent divergence dating was based on its capacity to coalesce complex evolutionary relationships. This tree served as a reliable backbone for divergence dating analysis performed using BEAST2 (Bouckaert et al. 2019), which allowed us to estimate divergence times with the incorporation of fossil calibration points for all 72 taxa included in our study. The integration of these methods ensured a comprehensive understanding of Conus evolution, with each strategy contributing unique strengths to the overall analysis.
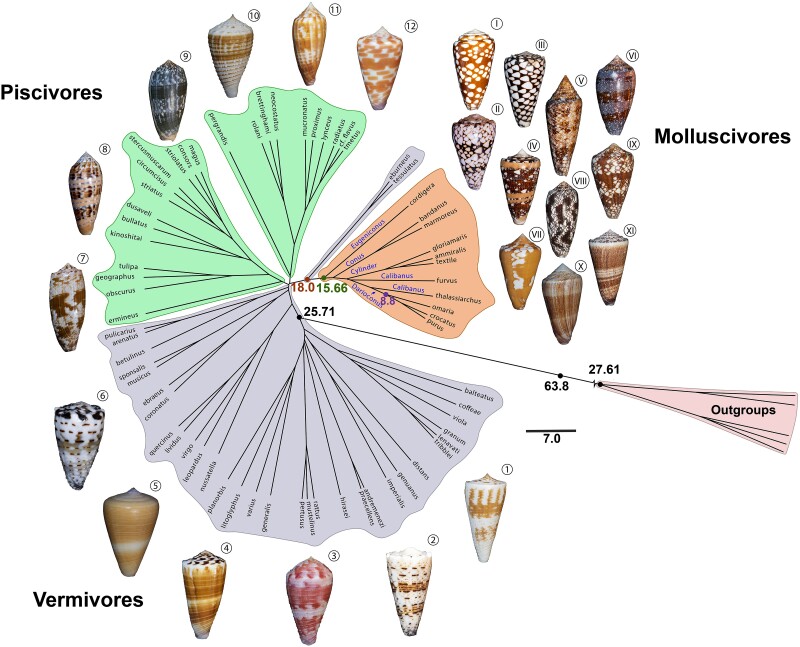
Phylogenetic reconstructions of 67 Conus species (11 molluscivores, 23 piscivores, and 33 vermivores) and 5 Conasprella species (outgroup), utilizing 86 HKG's derived from transcriptomic analysis, support a single transition to molluscivory within Conus (Fig. 1, supplementary figs. S1 to S4, Supplementary Material online). Divergence dating analyses revealed that the crown-group Conus evolved ∼25.7 Ma (95% highest posterior density [HPD]: 22.35 to 28.02 My) during the late-Oligocene (Fig. 1, supplementary fig. S4, Supplementary Material online). Furthermore, a compressed backbone highlights that most crown groups within Conus originated in the past 20 My. Compared with the other feeding groups, molluscivores are the most recently evolved. The clade of mollusk-hunting snails diverged from its sister lineage Tesselliconus, a group of vermivorous cone snails (Puillandre et al. 2014; Fig. 1) in the Early to Middle Miocene (∼18 Ma; HPD 16.36 to 19.67 My). Within the clade of molluscivorous snails, the branch leading to Eugeniconus (represented by Conus cordigera) was recovered as the oldest lineage, diverging from the remaining mollusk hunters ∼15.6 Ma (HPD 14.22 to 17.12 My), while Darioconus was recovered as the youngest lineage, diverging from Calibanus ∼8.8 Ma (HPD 8.03 to 9.75 My; Fig. 1, supplementary fig. S4, Supplementary Material online).
Fig. 1.
Reconstruction of Conus evolutionary history, highlighting the different feeding groups and divergence timing analysis. Multilocus-derived multispecies divergence timing analysis tree to display the taxa analyzed in this study indicating a single evolution of molluscivory. Feeding groups are highlighted: gray (vermivores), green (piscivores), and orange (molluscivores). Colored circles indicate estimated divergence times for nodes of interest (see supplementary fig. S4, Supplementary Material online for credibility intervals) obtained with BEAST using species coalescent topology generated with ASTRAL-III as input tree (see supplementary fig. S3, Supplementary Material online for ASTRAL tree). Values shown are in millions of years. Support values for all the nodes are found in supplementary fig. S3, Supplementary Material online (see also supplementary figs. S1 and S2, Supplementary Material online). The node corresponding to the ancestor of Tesseliconus and the mollusk-hunting species was recovered with maximum support. Outgroup refers to Conasprella species (see supplementary figs. S1 and S2, Supplementary Material online for additional information). Worm and fish-hunting Conus species represented in the picture are: (1) C. tribblei; (2) C. imperialis; (3) C. pertusus; (4) C. planorbis; (5) C. quercinus; (6) C. musicus; (7) C. geographus; (8) C. dusaveli; (9) C. striatus; (10) C. rolani; (11) C. lynceus; (12) C. tessulatus. Mollusk hunters are as follows: (I) C. cordigera; (II) C. bandanus; (III) C. marmoreus; (IV) C. ammiralis; (V) C. gloriamaris; (VI) C. textile; (VII) C. crocatus; (VIII) C. omaria; (IX) C. purus; (X) C. furvus; (XI) C. thalassiarchus.
The evolutionary shift from vermivory to molluscivory likely entailed genetic and molecular innovations specifically tailored to a novel prey type. We hypothesize that some of these evolutionary innovations would manifest as novel conotoxins exclusively expressed by molluscivorous Conus. One such component could be the χ-conotoxins. Currently, no χ-conotoxins have been reliably purified or sequenced from other Conus feeding groups, suggesting that they may be exclusive to molluscivores. χ-Conotoxins are the only known conopeptide that targets a transporter (Sharpe et al. 2001; Espiritu et al. 2023).
Molluscivores Express a Distinct Class of T-Superfamily Conopeptides
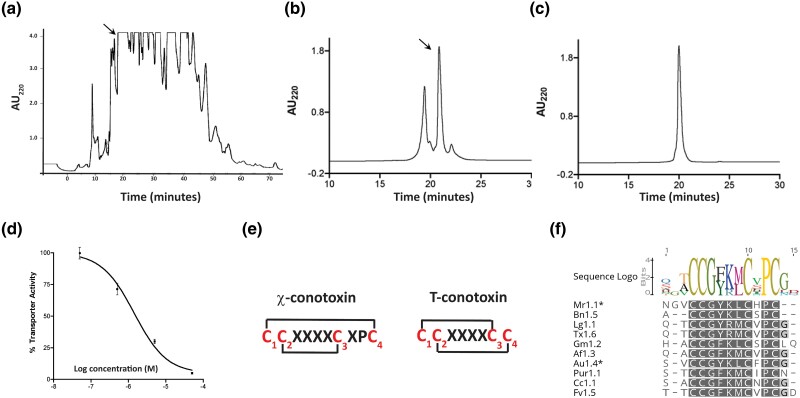
Since χ-conotoxins inhibit the human norepinephrine transporter (hNET), we adapted an assay (Galli et al. 1995) to screen Conus venoms for inhibitory activity against hNET stably expressed in human embryonic kidney (HEK 293) cells. Using this assay, we purified a putative χ-conotoxin from Conus aulicus, a species in the Darioconus clade (Fig. 2a to c). Venom from C. aulicus was extracted from pooled venom ducts; thus, it is not clear whether this is for defensive or predatory purpose. N-terminal Edman sequencing resulted in the sequence SVCCGYKLCFOC-NH2 (O: hydroxyproline). This sequence is similar to χ-MrIa (Table 1, supplementary tables S1, S3 to S5, Supplementary Material online). The synthetic peptide with a disulfide bridge between Cys 1 and Cys 4 and between Cys 2 and Cys 3 co-eluted with the native peptide (supplementary fig. S5, Supplementary Material online) and inhibited the reuptake of norepinephrine via hNET (IC50 of 2.6 ± 0.4 μM; n = 3 independent experiments; Fig. 2d). Evidence from the high-performance liquid chromatography (HPLC) experiments (Fig. 2a to c) suggested that this peptide is a major component of the C. aulicus venom. We propose to call this peptide χ-AuId following the standard nomenclature for conotoxins (Olivera and Cruz 2001).
Fig. 2.
χ-Conopeptide is highly expressed in the venom of C. aulicus and these peptides form a structurally and pharmacologically distinct family in the T-superfamily of conotoxins. a) HPLC chromatogram showing the absorbance profile at 220 nm of crude C. aulicus venom fractionated on a preparative C18 column using a linear gradient of 1.3% B90/min at a flow rate of 20 mL/min. The fraction marked by the arrow exhibited activity against hNET, as described in the Results section. b) Analytical HPLC chromatogram profile of the subfractionation of the active component in (a). The subfractionation was done using a linear gradient of 30% to 50% B90 for 20 min at a flow rate of 1 mL/min. The active peak is indicated by the arrow. Note that the hNET active fraction is the major component of this solution. c) Analytical HPLC chromatogram profile showing the homogeneity of the active fraction in (b). The HPLC experiment was done using the same linear gradient described in (b). The component that inhibits hNET has the sequence: SVCCGYKLCFOC# (O = hydroxyproline, # = C-terminal amidation). This purified fraction is designated as χ-AuId. d) Dose-dependent inhibition of hNET by χ-AuId. IC50 value of χ-AuId against hNET is 2.6 ± 0.4 μM. This value is the average from three experiments done in triplicate. e) Comparison of the cysteine framework between χ-conotoxins and canonical T-superfamily peptides. f) Sequence logo of χ-conotoxins. Conservative substitutions in amino acids between Cys 2 and Cys 3 are observed in these peptides. The sequences are derived from Mr1.1: C. marmoreus; Bn1.5: C. bandanus; Lg1.1: C. legatus; Tx1.6: C. textile; Af1.3: C. ammiralis; Au1.4: C. aulicus; Pur1.1: C. purus; Cc1.1: C. crocatus; and Fv1.5: C. furvus.
Table 1.
χ-Conotoxins are expressed by all species analyzed, representing five clades of molluscivorous Conus. These peptides define a distinct pharmacological family in the T-superfamily.
| Species | Peptide | Sequence | Subgenus (Clade) | |
|---|---|---|---|---|
| χ-conotoxins | C. marmoreus | Mr1.1a | NGVCCGYKLCHPC–– | Conus |
| C. bandanus | Bn1.5 | ––ACCGYKLCSPC- | Conus | |
| C. legatus | Lg1.1 | -QTCCGYRMCVPCG- | Cylinder | |
| C. textile | Tx1.6 | -QTCCGYRMCVPCG- | Cylinder | |
| C. gloriamaris | Gm1.2 | HACCGFKLCSPCLQ | Cylinder | |
| C. ammiralis | Af1.3 | -QACCGFKMCVPCG- | Cylinder | |
| C. aulicus | Au1.4b | SVCCGYKLCFPCG- | Darioconus | |
| C. purus | Pur1.1 | -STCCGFKMCIPCN- | Darioconus | |
| C. crocatus | Cc1.1 | -SACCGFKMCNPCG- | Darioconus | |
| C. furvus | Fv1.5 | -TTCCGFKMCVPCGD | Calibanus | |
| C. cordigera | Cdg1.2 | QLCCGYKLCW-C | Eugeniconus |
χ-MrIa (Balaji et al. 2000; McIntosh et al. 2000; Sharpe et al. 2001) and χ-AuId were purified from venom. GenBank accession numbers are in supplementary table S2, Supplementary Material online.
aχ-MrIa is encoded by clone Mr1.1.
bχ-AuId is encoded by clone Au1.4.
The next question we posed was whether peptides homologous to χ-AuId and χ-MrIa could be identified in the available venom gland transcriptome of snails representing five clades of molluscivores. Two groups of sequences with cysteine patterns CCXXXXCXXC and CCXXXXCXC were identified. The precursor sequences coding for these peptides displayed the canonical organization of conotoxin precursors (supplementary tables S3 and S4, Supplementary Material online), consisting of a highly conserved signal sequence, a relatively conserved propeptide, and a mature peptide region that in most cases is hypervariable. Sequence alignment with known T-superfamily conotoxins demonstrates that, like χ-MrIa, these peptides belong to the T-superfamily (supplementary table S4, Supplementary Material online). The mature peptide sequences in supplementary table S1, Supplementary Material online can be grouped into four classes. The largest class (Group 1), comprises 11 out of 20 sequences and includes Mr1.1, the clone that encodes for χ-MrIa, and Au1.4, the clone that encodes for χ-AuId (Table 1 and supplementary tables S1, S3, and S4, Supplementary Material online). Peptides in this group are designated as χ-conotoxins based on: (i) the inclusion of χ-MrIa and χ-AuId, the hNET inhibitors and (ii) conservative substitutions of amino acids between Cys2 and Cys3 were shown to maintain inhibitory activity against hNET, albeit changing the potency of the peptide against the transporter (Brust et al. 2009; Espiritu et al. 2023). GenBank accession numbers of these peptides are given in supplementary table S2, Supplementary Material online.
The χ-conotoxins are highly expressed in the mRNA transcript of all clades of molluscivorous Conus that we analyzed (Table 1, supplementary table S5, Supplementary Material online). These peptides have sequences with conservative substitutions in the first loop between Cys2 and Cys3 (consensus sequence: –Cys2-G(Y/F)(R/K)(L/I/M)-Cys3; Fig. 2f). The second loop has two amino acids and a conserved proline residue preceding Cys4 except for Cdg1.2 in which tryptophan is the only amino acid in the second loop. Figure 2e illustrates how the cysteine pattern of χ-conotoxins as well as the other peptides in supplementary table S1, Supplementary Material online diverge from the canonical T-superfamily conotoxin cysteine pattern (Balaji et al. 2000; Gupta et al. 2010). We propose that the division of the T-superfamily, to which these peptides belong, be called the 2-loop T conotoxins (2-loop T's) in contrast to the single intercysteine loop (Framework V) in the “standard” T-conotoxins (Walker et al. 1999; Terlau and Olivera 2004; Robinson and Norton 2014).
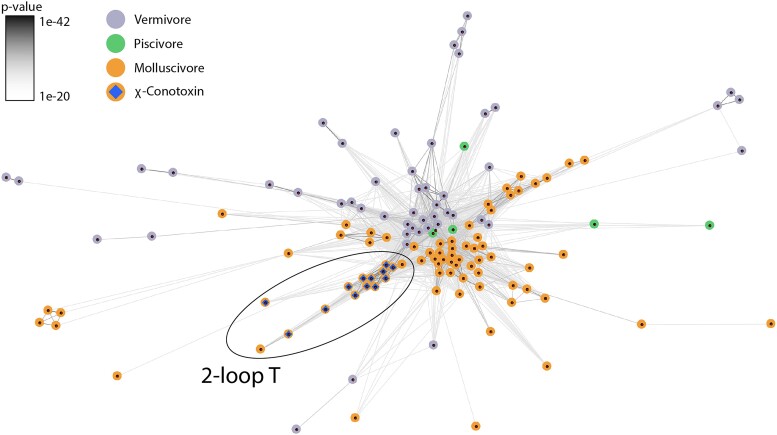
To investigate the evolutionary relationship of χ-conotoxins within the larger T-superfamily of conotoxins, we extracted T-superfamily sequences from a recent publication (Koch et al. 2024), which include toxins from 42 phylogenetically diverse cone snails with different dietary preferences. This dataset was procured by carefully remapping the raw reads back to the assembled transcripts. This process ensures that: (i) “hidden” isoforms that are easily filtered out by assembly programs, such as Trinity, are recaptured, and (ii) assemblies with limited support are removed. We performed a CLANS clustering analysis, which we have previously shown, can resolve the evolution of contoxins (Koch et al. 2022; Hackney et al. 2023). Briefly, this clustering approach used all-against-all blastp P-values as attractive forces in a uniformly repulsive field, which will cause sequences of high similarity to attract one another and form clusters. We found that the 2-loop T-conotoxins and its subgroup the χ-conotoxins are exclusively found in molluscivores and belong to a single lineage (Fig. 3). These toxins belong to a distinct subgroup in the T-superfamily conotoxins from vermivores, piscivores, and molluscivores. A maximum likelihood gene tree of the sequences belonging to this subgroup further supports a single origin of χ-conotoxins and demonstrates that χ-conotoxins evolved in molluscivores (supplementary fig. S6, Supplementary Material online). The adaptive branch-site method aBSREL (Smith et al. 2015) found no evidence of episodic diversifying evolution on the branch leading to the 2-loop T/χ-conotoxins.
Fig. 3.
BLOSUM62 CLANS cluster map of χ-conotoxins and other T-superfamily toxins. Colored circles represent individual toxin precursor sequences, and the connecting edges show the blastp P-values above 1e−20 between the connected circles. The circles are color coded according to prey preferences (gray: vermivores, green: piscivores, orange: molluscivores). The χ-conotoxins (represented by a blue prism) are all part of a lineage of 2-loop Ts, which are exclusively found in molluscivores.
Effects of χ-Conotoxins in Mollusks
χ-Conotoxins are expressed in all clades of molluscivorous cone snails that we analyzed. Further, our transcriptomic analysis failed to identify χ-conotoxins outside of the molluscivorous clades (Fig. 3, supplementary fig. S6, Supplementary Material online). These data suggest that χ-conotoxins serve an important role in the hunting for mollusks and may alter prey behavior to facilitate prey capture.
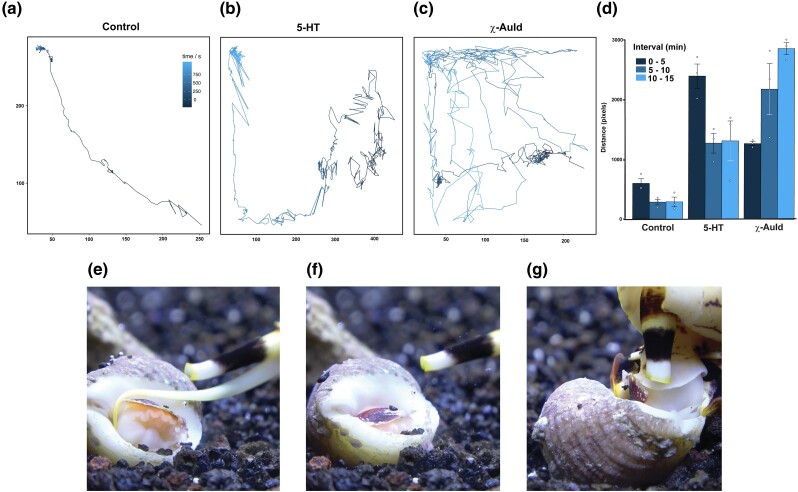
To test this, we injected χ-AuId into the model organism Aplysia californica, a gastropod mollusk whose behavior has been well characterized (Mackey and Carew 1983; Leonard and Lukowiak 1986). Injection of 10 nmol χ-AuId resulted in increased locomotion compared with the control (Fig. 4a, c, and d). Distinctive symptoms occurred ∼5 min postinjection (see Fig. 4d); the animals failed to gain traction while locomoting as evidenced by the inability of parts of the foot to stick to the tank walls. This symptomatology progressed to a point where the entire foot was detached from the tank wall, and the animal fell. Impaired locomotion was coupled with random hyperactive movement without any clear direction. This behavioral effect is dose-dependent as 1 nmol of χ-AuId neither resulted in increased locomotion nor behavioral phenotype significantly different from the control, within the 15-min observation period (supplementary fig. S7, Supplementary Material online).
Fig. 4.
Behavioral phenotype after injecting χ-AuId into A. californica and images of C. bandanus envenomation progression. Video traces of the Aplysia body position showing the path traveled over a 15 min period after injection with: a) saline control, b) 0.01 nmol serotonin (5-HT), and c) 10 nmol χ-AuId. d) Quantification of the distance traveled (in number of pixels). The bar for each treatment (n = 3, open circles) represents the distance traveled over a 5 min period (see inset). The P-values are: χ-AuId (0 to 5 min, P = 0.0067; 5 to 10 min, P = 0.0459; 10 to 15 min, P = 5.7E-5), 5-HT (0 to 5 min, P = 0.006; 5 to 10 min, P = 0.02; 10 to 15 min, P = 0.084). Error bars represent standard deviation. e) to g) series of photographs showing events of C. bandanus envenomation of a prey snail. e) Conus bandanus envenomates prey. f) Prey tries to withdraw into the shell (note the operculum almost sealing off the shell). g) Minutes later, the prey emerges straight into the rostrum (false mouth) of C. bandanus.
Our data and previously published works (Sharpe et al. 2001; Espiritu et al. 2023) demonstrate that χ-conotoxins inhibit hNET. The prey of molluscivorous cone snails, however, does not appear to functionally express NET or the related octopamine transporter (Adamo 2008). A BLAST search using the hNET against the A. californica genome showed that the closest matches are the A. californica serotonin transporter (aSERT) and a sequence predicted to be a norepinephrine transporter (aNET). SERT and NET are closely related and belong to the same branch of the solute carrier 6 family of Na+/Cl− dependent transporters (Broer and Gether 2012). It was shown that Leu469 in hNET is important for χ-MrIa activity and substituting this with Phe from an equivalent position in the human dopamine transporter severely reduces activity in the L469F mutant (Paczkowski et al. 2007). Sequence alignment of hNET with aSERT and with the putative aNET showed that Ile occupies the position equivalent to L469 in aSERT, and the same position is occupied by Phe in the putative aNET (supplementary fig. S8, Supplementary Material online). The conservative substitution of Leu469 in hNET with Ile in aSERT suggests a possibility that χ-conotoxins may inhibit aSERT.
SERT is the major mechanism terminating serotonergic neurotransmission by the reuptake of serotonin back to the presynaptic terminal (Blakely et al. 1991). Blocking the transporter results in increased synaptic serotonin (5-HT) leading to a prolonged activation of post-synaptic receptors. If χ-conotoxins perturb serotonergic neurotransmission presumably by inhibiting aSERT, then a hyperlocomotive phenotype could also result from a transient spike in serotonin resulting from the injection of the neurotransmitter into A. californica. Injection of 0.01 nmol serotonin resulted in an immediate hyperlocomotive phenotype (Fig. 4a, b, and d). Although A. californica became hyperlocomotive, there is a notable inability of the animal's foot to completely stick to the tank wall, resulting in impaired locomotion—a behavior similar to χ-AuId injection into A. californica. A 0.001 nmol injection of serotonin and a 0.01 nmol injection of octopamine did not result in behavior significantly different from the saline-injected control (supplementary fig. S7, Supplementary Material online).
Interestingly, behavior similar to that of A. californica upon injection with χ-AuId was observed when Conus bandanus envenomated a prey snail. Conus bandanus is one of the molluscivores that expresses transcripts for χ-conotoxin (Table 1, supplementary tables S1, S3, and S5, Supplementary Material online). Upon envenomation, the prey appeared to withdraw into the shell; however, it did not remain inside for long. The prey snail came out of its shell and exhibited disorganized squirming and twitching. This behavior continued to a point where the foot was extended out of the shell in a presumed attempt to crawl. Unfortunately, for the prey, the foot went straight into the mouth of C. bandanus (Fig. 4e to g, supplementary video S1, Supplementary Material online). In a separate C. bandanus envenomation event, two different prey species were able to escape the initial strike, both appeared to become hyperlocomotive but were repeatedly falling from the tank walls; behaviors that are very similar to that of A. californica when injected with χ-AuId (supplementary videos S2 and S3, Supplementary Material online).
Discussion
The phylogenetic reconstruction presented in this study aligns well with previous Conus phylogenies (Abalde et al. 2019; Lee and Park 2022). Our species tree analysis, expanding on molluscivores, confirms the monophyly of molluscivory in Conus, consistent with prior studies (Espiritu et al. 2001; Puillandre et al. 2014; Uribe et al. 2017; Abalde et al. 2019; Lee and Park 2022; Koch et al. 2024). However, our study provides enhanced resolution within this clade, particularly in pinpointing the divergence timing and evolutionary transitions specific to molluscivory. By incorporating novel transcriptomic data, our study sheds light on genetic adaptations, such as the emergence of χ-conotoxins, that were not the primary focus of earlier reconstructions. While previous studies have provided a broad understanding of Conus evolution, our detailed approach offers a deeper exploration of the genetic innovations associated with the molluscivorous lineage, highlighting the evolutionary processes that have shaped this unique group within Conus. Here, we show that the genus Conus underwent rapid diversification in the Miocene and the lineage of molluscivorous Conus first appeared in this geologic epoch.
The Miocene was a crucial time for the diversification of the Indo-Pacific lineages of cone snails. Of the three major dietary groups in Conus (vermivores, piscivores, and molluscivores), molluscivores are the least diverse group, constituting <10% of all known Conus species (Puillandre et al. 2014). As shown here, molluscivory evolved once in Conus ∼18 Ma in the Early Miocene. The Early Miocene is characterized by major tectonic and oceanic events. The Indian subcontinent had pushed far into Eurasia uplifting the Tibetan plateau, while the Tethys Seaway linking the Atlantic and the Indian Oceans was increasingly restricted (Bradshaw 2021). The tectonic collision of the Australian plate with SE Eurasia (∼20 to 25 Ma) led to an increase in coral-carbonate platforms (Williams and Duda 2008). Together, the narrowing of the Tethys Seaway and the collision of the Australian plate with SE Eurasia are probably linked to an increase in habitat complexity and availability, factors that might have contributed to Conus diversification (Williams and Duda 2008). Our analyses corroborate an Oligocene–Miocene burst of diversification for the cone snails, with several Conus lineages originating during that time (see Fig. 1 and supplementary fig. S4, Supplementary Material online). It is possible that a transition from vermivory to molluscivory was facilitated once new niches became available in response to geological events that occurred during the Early Miocene. Such a prey shift likely involved modification and repurposing of the arsenal of venom peptides from the ancestral state (presumably adapted to polychaete worm prey) to effectively target a new prey type. In principle, this process explains the observations that are documented above for the χ-conotoxins.
The T-superfamily is generally expressed across the entire genus Conus (Walker et al. 1999), but the subset of χ-conotoxins appears to be predominantly expressed in the molluscivorous lineage. This suggests that the switch to a molluscivorous diet was accompanied by molecular evolution in the T-superfamily giving rise to the χ-conotoxins. χ-Conotoxins are among the most highly expressed gene products in the venom gland transcriptome of C. aulicus. The high expression level of the χ-AuId transcript in the C. aulicus transcriptome corresponds with high abundance in the venom, as χ-AuId appears as a major peak in the HPLC fractionation of the crude C. aulicus venom. The high transcript expression levels and peptide abundance are consistent with these peptides playing an important role in predation. Injection of χ-AuId into A. californica provides support for this assertion. A 10 nmol injection of χ-AuId induced impaired movement manifested by the inability to locomote, escape, and adhere to the substrate. χ-AuId resulted in a dramatic, uncoordinated hyperactivity with the snail constantly climbing the walls of the aquarium but quickly falling off. The observed behaviors in A. californica suggest a role for χ-conotoxins in addressing common challenges faced by molluscivorous Conus.
When gastropods face physiological challenges, molluscivorous predators encounter a dilemma: their prey's reaction is to retreat deeply into their sturdy shells for protection. Most successful specialized predators of gastropods have evolved mechanisms for drilling a hole or breaking the shell. Our results show that molluscivorous Conus has evolved a pharmacological agent that prevents prey from withdrawing deep into their shells. The uncoordinated hyperactivity induced by χ-AuId could lead envenomated prey to ultimately emerge from the shell, completely defenseless. Observations of molluscivorous cone snails maintained in laboratory aquaria showed that these predators envenomate their prey multiple times. Thus, a hyperactive snail with impaired locomotion is vulnerable to further envenomation attacks, allowing the predatory Conus to deliver more venom, effectively subduing their prey. A video of C. bandanus (supplementary video S1, Supplementary Material online), a molluscivorous Conus from which we have identified a χ-conotoxin in its transcriptome, shows that minutes after envenomation, the prey snail emerges from within the shell, allowing the C. bandanus to engulf the prey, ultimately extracting all the soft tissue. Since the same problem is faced by any cone snail that specializes in gastropods as prey, the original shift to molluscivory may have been accompanied by strong selection for the evolution within the T-superfamily of χ-conotoxin bioactivity. There would also be selective pressure to retain expression of a χ-conotoxin in venom as the different lineages of molluscivorous cone snails diverged from the original ancestral molluscivore. We postulate that the ancestral species that gave rise to molluscivory had a χ-conotoxin-like component in its venom that was very similar in structure and sequence features to the χ-conotoxins presented here. This ancestral peptide could serve either predatory or defensive functions since the predator and prey of mollusk-hunting snails are primarily other mollusks. Once this branch of the T-superfamily was established, opportunities to repurpose the basic structural motif of χ-conotoxins for other functional purposes may have occurred, as can be seen in the Groups 2 to 4 peptides shown in supplementary table S1, Supplementary Material online.
Although we did not find χ-conotoxin transcripts outside of the molluscivores and provided evidence that this peptide family evolved in mollusk hunters, other groups have reported χ-conotoxins from the fish-hunting Conus magus (Pardos-Blas et al. 2019) and Conus striatus (cDNA sequence; Wu et al. 2013) and the worm-hunting Conus ebraeus (Pardos-Blas et al. 2022). However, a recent work (Koch et al. 2024) failed to identify χ-conotoxin transcripts outside the mollusk-hunting group. Consistent with this, our analyses of T-superfamily transcripts, including those from C. magus, C. striatus, and C. ebraeus showed that χ-conotoxins are expressed only in the molluscivorous group. Moreover, a maximum likelihood gene tree of the sequences belonging to the T-superfamily further supports a single origin of χ-conotoxins and demonstrates that χ-conotoxins evolved in molluscivores. In support of these, χ-conotoxins were not found in the C. ebraeus (894.86 tpm in one specimen; Pardos-Blas et al. 2022) and C. magus milked predatory venom (Rogalski et al. 2023) proteomes, whereas this work and others showed χ-conotoxins are expressed in the venom of molluscivorous Conus (Balaji et al. 2000; McIntosh et al. 2000; Sharpe et al. 2001; Gupta et al. 2010; Dutertre et al. 2013, 2014; Abalde et al. 2021; Espiritu et al. 2023; Ratibou et al. 2024). In contrast to the very high expression of χ-conotoxins in molluscivores, these peptides are expressed at very low tpm values in nonmollusk-hunting snails except for one specimen of C. ebraeus (see supplementary table S5, Supplementary Material online). Different specimens of the same species in the nonmollusk-hunting snails express different χ-conotoxin transcripts, whereas different specimens of the same species in molluscivores, if available, express the same χ-conotoxins (see supplementary table S5, Supplementary Material online). Based on these observations, it is likely that χ-conotoxins identified in nonmollusk-hunting snails originate from exogenous sources. The evidence, we present here, indicates a single origin of χ-conotoxins and the broader 2-loop T-conotoxins in molluscivorous Conus. Additional support for this could be obtained from a synteny-based analysis which may be possible once genomes from molluscivorous Conus are available.
Previous work has shown that shifts in prey preference are the main determinants of venom composition in cone snails (Koch et al. 2024). Principal component analysis of conotoxin transcripts from 42 Conus species representing all feeding groups showed clear separation of each group (Koch et al. 2024). Thus, each feeding group appears to have a distinct repertoire of peptides in their venom tailored specifically to perturb behavioral circuits in their prey. The evidence we provide here supports the idea that χ-conotoxins and the larger 2-loop T-conotoxins are exclusively expressed in molluscivores and have evolved specifically within this group. The effects of χ-conotoxins on the behavior of the mollusk A. californica that we present here offer a possible rationale for the exclusive expression of this toxin among the mollusivores. However, it cannot be ruled out that venom components from worm or fish-hunting snails might induce the same effect and target the same circuit in A. californica as the χ-conotoxins. In the wild, however, this is very unlikely to be observed since these snails are specialist hunters of worms or fish (Kohn 1959; Rockel et al. 1995; Terlau and Olivera 2004).
In this paper, we report a possible role of χ-conotoxins in the prey capture of mollusk-hunting cone snails. Previous work reported that χ-MrIa from Conus marmoreus serves predatory functions (Dutertre et al. 2013), while another publication showed that the peptide is a component of the defensive venom cocktail used by the snail to deter predators (Dutertre et al. 2014). It should be noted that the defensive venom was induced immediately after the first predatory strike; however, mollusk-hunting cone snails envenomate prey multiple times. Thus, an alternative interpretation of the data is that the venom released in the putative defensive stinging incident following the initial predatory strike is a part of a series of predatory venom injections—the endpoint of which is to take down prey. Since both the predator and prey of molluscivorous Conus are typically other mollusks, the distinction between defensive and predatory venom components becomes unclear. For mollusk-hunting cone snails, venom components, such as the χ-conotoxins, can serve both predatory and defensive purposes (Abalde et al. 2021; Ratibou et al. 2024).
Here, we show that χ-conotoxins affect locomotion in Aplysia californica. A. californica locomotion is under the control of the neurotransmitter serotonin (Mackey and Carew 1983; Marinesco et al. 2004; Gillette 2006; Aonuma et al. 2020). A BLAST search using the human NET sequence against the A. californica genome showed that one of the closest matches is the A. californica serotonin transporter (aSERT). Importantly, sequence alignment showed that Leu469 in hNET which was shown to be important for χ-MrIa activity (Paczkowski et al. 2007) is substituted by Ile in an equivalent position in aSERT (see supplementary fig. S8, Supplementary Material online). This conservative substitution likely maintains χ-conotoxin activity against aSERT. SERT is the major mechanism that terminates serotonergic signaling via the reuptake of serotonin back into the post-synaptic terminal (Blakely et al. 1991). Serotonin injection produced phenotypes similar to that of χ-AuId, while the same amount of octopamine, a molluscan neutrotransmitter, structurally similar to norepinephrine, did not alter A. californica behavior significantly different from control (see supplementary fig. S7, Supplementary Material online), results consistent with previously published work (Mackey and Carew 1983). The similarity of behavioral effects from both χ-AuId and serotonin indicates that these compounds affect the same circuit in A. californica. Thus, our data suggest that χ-conotoxins affect serotonergic circuits, likely via inhibition of aSERT. This can be rationalized from the temporal differences in the onset of effects between χ-AuId and serotonin. Since χ-AuId inhibits aSERT, time is required for the neurotransmitter to reach an effective concentration to act on postsynaptic receptors and produce the behavior observed, whereas the same concentration is reached faster from serotonin injection. Thus, the onset of behavioral phenotype is observed within 1 min of serotonin injection, while χ-AuId induces behavioral effects ∼5 min postinjection (see Fig. 4d).
From the data presented here, we conclude that χ-conotoxin is one of the biochemical innovations of molluscivorous cone snails adapted to address biological problems unique to mollusk hunting. Since the major predators of these snails are other mollusks, these peptides can easily be adapted for defense. We propose that χ-conotoxin-induced uncoordinated hyperactivity solves the biological problem posed by the shell. Hyperactive snails consistently emerge from their shells, allowing the predator to engulf their prey or deliver more venom, as shown here when C. bandanus envenomated its prey. Impaired locomotion resulting from the activity of the peptide prevents escape which gives an additional advantage to the molluscivorous Conus. Although χ-conotoxin is sufficient to phenocopy the observed behavior in prey upon envenomation, it is highly possible that other components in venom could target a different component of the locomotory circuit in prey producing the same phenotype as the χ-conotoxins. In combination, the activity of these peptides might lead to a faster onset of symptoms, resulting in a more successful predation attempt. The synergistic activity of different conotoxins to induce rapid immobilization of prey has been postulated for piscivorous snails (Olivera 1997). The data presented here highlights the usefulness of Conus as a model for understanding how ecological pressure and the corresponding genetic and molecular adaptations allow species to diversify and colonize novel ecological niches.
Materials and Methods
Transcriptome Analysis
Sequences from transcriptome libraries of molluscivorous cone snails identified as potential T-superfamily conotoxin sequences were scrutinized for the characteristic organization of conopeptide precursors: a highly conserved N-terminal signal sequence, followed by a more variable propeptide region, and a hypervariable mature peptide region at the C-terminus sharing the typical arrangement of Cys residues (Frameworks I and V) associated with χ- and T-conotoxins, respectively. Toxin sequences were aligned using MacVector 17.0 sequence analysis software (MacVector, Inc., Apex, NC, USA) and refined by eye. The sequences described in this publication have been deposited into GenBank (supplementary table S2, Supplementary Material online).
Venom gland transcriptomes were generated, as previously described (Li et al. 2018). Briefly, adapter clipping and quality trimming of raw reads were performed using fqtrim software (version 0.9.4, http://ccb.jhu.edu/software/fqtrim/) and PRINSEQ (version 0.20.4; Schmieder and Edwards 2011). After processing, sequences <70 bp and those containing >5% ambiguous bases (Ns) were discarded. De novo transcriptome assembly was performed using Trinity Version 2.0.5 (Altschul et al. 1990; Grabherr et al. 2011) with a k-mer size for building De Bruijn Graphs of 31, a minimum k-mer coverage of 10, and a minimum glue of 10. Assembled transcripts were annotated using Blastx (NCBI-Blast-2.2.28+; Altschul et al. 1990) against an in-house database compiled from ConoServer (Kaas et al. 2008) and UniProtKB/Swiss-Prot (RRID:SCR_004426; UniProt Consortium 2015) protein sequence databases. Common genes that were shared between all datasets were identified using the blast identities (e < 10−4) of assembled contigs. See supplementary figs. S1 to S3, Supplementary Material online, for the species analyzed for HKG transcript expression.
Phylogenetic Analyses
We aligned each locus (available online) using the program MAFFT v7.130b (Katoh and Standley 2013) with optimized parameters for each gene. We then trimmed poorly aligned regions and gap-only columns using the program GBlocks v.0.91b (Talavera and Castresana 2007), with reduced stringency parameters (b1:0.5, b2:0.5, b3:12, b4:7). We then concatenated all loci into a supermatrix and inferred a preliminary tree using IQ-Tree v2.1.3 (Minh et al. 2020) and carried out multiple phylogenetic reconstructions to explore the effects of data partitioning. We examined two different partitioning schemes: unpartitioned and partitioned by locus. For both schemes, we inferred phylogenetic relationships of Conus species with IQ-Tree v2.1.3 (Minh et al. 2020), and assessed branch support by performing 2,000 replicates of the ultrafast bootstrap approximation (UFB; Hoang et al. 2018) and 2,000 replicates of the branch-based, SH-like approximate likelihood ratio test (Guindon et al. 2010). For the unpartitioned dataset, we analyzed the supermatrix with GTR + G as the model of nucleotide evolution. For the partitioning by locus, we partitioned the supermatrix by locus and used the partition file to run ModelFinder2 (Kalyaanamoorthy et al. 2017) within IQ-Tree v2.1.3. For the ModelFinder2 analysis, we used the AICc model selection criterion. Using the partitioning scheme, we inferred phylogenetic relationships by selecting the “—-p” option for partitioning and the best model of sequence evolution recovered by ModelFinder2 for each locus, and assessed branch support by performing 2,000 UFB replicates.
We quantified genealogical concordance in our topology for the partitioned by locus analysis with the coalescent program ASTRAL-III (Zhang et al. 2018) to account for gene tree discordance arising from independent lineage sorting while inferring a species tree. For the ASTRAL-III analyses, we first estimated individual gene trees with IQ-Tree, using model selection for each locus and 2,000 UFB replicates. We then combined all output trees into a single file, collapsed all nodes with ≤30% UFB support using Newick Utilities (Junier and Zdobnov 2010), and estimated the species tree using ASTRAL-III (Zhang et al. 2018). Support was assessed using the default local posterior probability. A list of HKGs and files used for phylogenetic analysis can be found in figshare (https://doi.org/10.6084/m9.figshare.26875987.v1).
Divergence Dating
We inferred a dated time tree for Conus and its species using BEAST2 v2.6.3 (Bouckaert et al. 2019). We selected two secondary calibrations obtained from Abalde et al. (Abalde et al. 2019) to calibrate the analyses: the crown group divergence age of Conidae (mean set to 62 Ma, 95% confidence interval set to 3.0 Ma) to calibrate the root (normal calibration), and Conasprella, sister taxon to Conus (mean set to 44.25 Ma, 95% confidence interval set to 5.4 Ma) as an additional calibration (normal calibration). For our BEAST2 runs, we used the complete dataset of 86 loci. For the input tree, we constrained the topology following our ASTRAL-III output, rooted on Conasprella. We then transformed the input tree to ultrametric with chronos in the R package “ape” (Paradis et al. 2004), a function that estimates the node ages of a tree using a semi-parametric method based on penalized likelihood (Sanderson 2002). The ultrametric tree was used for our subsequent analysis. The BEAST2 analyses were run on a single supermatrix without partitioning. We selected the HKY model of sequence evolution, with estimated frequencies and a strict clock model. We used a Yule model for the tree prior and turned off all tree topology search parameters. For each run, we set the chain length to 30 million generations, the sampling density to every 10,000 generations, and burn-in at 10%. We performed a total of three independent runs and assessed convergence across runs and parameter ESS values (>200) using the program Tracer v1.7 (Rambaut et al. 2018). Chronograms were summarized using the program TreeAnnotator v2.6.3 (Bouckaert et al. 2019). BEAST files are available through figshare (https://doi.org/10.6084/m9.figshare.26875984.v1).
Clustering Analysis
We extracted T-superfamily toxin precursor sequences from a recent paper (Koch et al. 2024) of 42 phylogenetically diverse cone snails with different prey preferences. We appended the χ-conotoxins described here that were not already present in the file. Using these sequences, we did CLANS clustering. This method calculates all-against-all blast P-values using the BLOSUM62 matrix and uses the negative log of these values as attractive forces in a uniformly repulsive field. This will cause sequences with high similarity to cluster together. We used a blast P-value cutoff at 1e−20, which identified a smaller cluster that contains the χ-conotoxins. Sequences used for the analysis is available through figshare (https://doi.org/10.6084/m9.figshare.26876092.v1).
Phylogenetic and Selection Analysis of T-Superfamily Conotoxins
Using the sequences from the χ-conotoxin containing cluster identified earlier, we performed a maximum likelihood gene tree reconstruction. The sequences were aligned using mafft v7.525 (Katoh and Standley 2013) using linsi settings. Using the PAL2NAL program (Suyama et al. 2006), we obtained a corresponding nucleotide alignment. The tree was generated using IQ-tree v2.2.2.3 (Minh et al. 2020) with seed 837,022 and the GTR + I + Gmodel of nucleotide evolution based on the built-in model selection module. The UF-boot was used with 1,000 replicates. A web version of aBSREL (https://datamonkey.org/absrel; Smith et al. 2015) was used to determine whether a proportion of sites evolved under positive selection. The nucleotide alignment used for the gene tree was uploaded to figshare (https://doi.org/10.6084/m9.figshare.26876092.v1).
Extraction and Fractionation of Crude Venom
Lyophilized venom (550 mg) from C. aulicus collected in the Philippines was extracted with 40 mL of an aqueous solution containing 40% acetonitrile (ACN) and 0.1% trifluoroacetic acid (TFA). The suspension was vortexed for 1 min and homogenized in a 50 mL glass-teflon homogenizer. The homogenate was vortexed for 10 s and centrifuged at 15,000 × g for 10 min. The supernatant was diluted with a solution containing 0.1% TFA and applied on a Vydac preparative reversed-phase C18 HPLC column (22 × 250 mm, 10 to 15 μm, 300 Å). The components of the venom were eluted using a linear gradient of buffers consisting of 0.1% TFA in water (Buffer A) and 0.1% TFA in 90% aqueous ACN (Buffer B90). The elution gradient was 1.3% B90/min at a flow rate of 20 mL/min. The absorbance of the eluent was monitored at 220 and 280 nm.
Purification of Conotoxins From C. aulicus
Peptides from C. aulicus were further purified from the corresponding fractions by analytical reversed-phase HPLC using a Vydac C18 monomeric column (4.6 × 250 mm, 5 μm, 300 Å). Elution of the components of the hNET-inhibiting fraction from the C. aulicus venom was done using a linear gradient of 30% to 50% buffer B90 for 20 min at a flow rate of 1 mL/min. The homogeneity of the fractions was verified by analytical HPLC using the same elution profile previously described.
Reduction and Alkylation of the Purified Peptides
For sequencing, the purified peptide from C. aulicus that showed inhibitory activity against the hNET was reduced using 1,4-dithiothreitol (DTT) and alkylated using 4-vinylpyridine (4-VP). The pH of the solution containing the peptide was buffered to pH 8 using 0.5 M Tris base. Disulfide bonds were reduced with 10 mM DTT at 65 °C for 30 min. After 30 min, 0.8 μL of pure 4-VP was added to the solution containing the reduced peptide. The alkylation of the reduced peptide was performed in the dark at room temperature for 30 min. The reduced and alkylated peptide was purified from the reaction using an analytical reversed-phase HPLC column (5 μm, 4.6 × 250 mm, 300 Å)
Peptide Sequencing
Approximately 70 pmol of the alkylated peptide was sequenced by Edman degradation at the DNA/peptide core facility, University of Utah. The 3-phenyl-2-thiohydantoin amino acid derivatives were identified by HPLC.
Mass Spectrometry
Matrix-assisted laser desorption ionization (MALDI) mass analysis was performed using the Applied Biosystems Voyager System 4,271 on reflector mode. The instrument was scanned over the m/z range between 0.7 and 3 kDa in positive ionization and in reflectron mode.
Peptide Synthesis
The peptide χ-AuId was synthesized with an Apex 396–automated peptide synthesizer using a standard solid-phase Fmoc (9-fluorenylmethyloxycarbonyl) protocol. χ-Au1d was synthesized on Fmoc-L-Cys(Acm)-Wang resin (substitution: 0.55 mmolg-1). All standard amino acids were purchased from AAPPTec and N-α-Fmoc-(O-tBu)-L-trans-4-hydroxyproline from NovaBiochem. Side-chain protection for the following amino acids was: Asp and Hyp: O-tert-butyl (O-tBu); Arg: 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl; Lys: tert-butyloxycarbonyl; Ser, Tyr, and Thr: tert-butyl (tBu); His: trityl (Trt). In order to control oxidative folding of the cysteines, orthogonal protection was used: one pair of Cys (Cys 1 and Cys 4) was Trt protected; the second pair (Cys 2 and Cys 3) was protected with acetamidomethyl group. Peptides were synthesized on 50 μmol scale. Coupling activation was achieved with one equivalent of 0.4 M benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate and two equivalents of 2 M N,N-diisopropylethyl amine in N-methyl-2-pyrrolidone (NMP) and 10-fold excess of amino acid. Each coupling reaction was conducted for 60 min. Fmoc deprotection reaction was carried out for 20 min with 20% piperidine in DMF.
Oxidative Folding of the Linear Peptides
The linear peptide was cleaved from 25 to 50 mg of resin using 1 mL reagent K (11.0 M TFA, 3.0 M 1,2-ethanedithiol, 4.0 M thioanisole, and 5.0 M phenol) and precipitated using ice-cold methyl tert-butyl ether (MTBE). After the removal of MTBE, the precipitate was resuspended in Buffer A (0.1% TFA) and purified using a Vydac C18 semipreparative reversed-phase column (5 μm, 10 × 250 mm, 300 Å) and using a linear gradient of 15% to 50% B90 for 35 min and a flow rate of 4 mL/min. Peptide folding was achieved in two steps. The first disulfide bridge was formed by incubating the linear peptide in a solution containing 1.0 mM oxidized glutathione, 2.0 mM reduced glutathione, 100 mM Tris (pH = 7.5) and 0.1 mM ethylenediaminetetraacetic acid for 2 h. The folded peptide was separated from the solution using a reversed-phase Vydac C18 semipreparative HPLC column (see column details and HPLC gradient above). The formation of the second disulfide bridge was achieved by incubating the peptide for 10 min in a solution containing 24% ACN (v/v), 380 mM TFA and 5 mM I2. The folded peptide was purified to at least 95% purity in the same manner as described above. The identity of the folded peptide was confirmed by mass spectrometry and co-elution with the native peptide isolated from venom. Briefly, a solution containing both native and synthetic peptides was loaded into a Vydac analytical HPLC column and eluted using the solvent gradient described above for the purification of the synthetic peptide, except that the flow rate was at 1 mL/min for analytical HPLC. Solutions containing either the native or synthetic peptides were also injected into an HPLC column and eluted in the same manner described previously. Retention times from the three HPLC runs were compared. Similarity in the retention times and, most importantly, a single HPLC peak resulting from the elution of the solution containing both the native and synthetic peptides are indicators of the successful folding of the synthetic peptide.
Cell Culture
Human embryonic kidney cells stably expressing the hNET (HEK293-hNET), a gift from the Blakely Lab (Vanderbilt University), were grown in Dulbecco's modified Eagle's medium containing 10% (v/v) FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine and 250 μg/mL G418 at 37 °C and 5% CO2 in a humidity controlled incubator.
Norepinephrine Uptake Assay
The norepinephrine uptake assay followed the protocol described previously (Galli et al. 1995). HEK293-hNET cells were plated at a density of ∼50,000 cells/well on amine coated 96-well tissue culture plates 1 d before the experiment. On the day of the experiment, the medium was aspirated and the cells washed once with Krebs-Ringer's-HEPES (KRH) buffer (130 mM NaCl, 1.3 mM KCl, 2.2 mM CaCl2, 1.2 mM MgSO4, 1.2 mM KH2PO4, 10 mM HEPES, 1.8 g/L dextrose, pH 7.4). The cells were preincubated with the conotoxins for 10 min at 37 °C in KRH buffer supplemented with 100 mM L-ascorbate, 100 mM pargyline, and 10 mM U-0521(catechol O-methyl transferase inhibitor). The uptake assay was initiated by the addition of norepinephrine, Levo-[ring-2,5,6-3H] to a final concentration of 50 nM. The uptake assay was performed at 37 °C for 15 min and terminated by rapid aspiration of the uptake buffer followed by 2 washes with 200 μL ice-cold KRH buffer. The cells were then lysed with 100 μL of a 10% SDS solution at room temperature with gentle shaking for 60 min. Accumulated radioactivity in the 50 μL of the lysate was measured using liquid scintillation spectrometry. The remaining 50 μL of the lysate was used to measure protein concentration using the DC protein assay kit (Bio-Rad), following the manufacturer's protocol. A parallel experiment was performed in the presence of 1.0 μM desipramine to account for nonspecific uptake.
Aplysia californica Bioassay
Juvenile A. californica (1 to 20 g) was purchased from the National Resource for Aplysia, University of Miami. Snails were acclimated to the testing conditions for at least 1 d prior to the experiment. Peptides were resuspended in sterile normal saline solution (NSS) and stored overnight at 4 °C. On the day of the experiment, snails were transferred to smaller observation tanks, and individual snails were injected near the anterior junction of the parapodia with 20 μL solution of the test compounds. Control animals were injected with the same volume of NSS. Test animals were injected with either 1 or 10 nmol χ-AuId, 0.001 and 0.01 nmol serotonin, and 0.01 nmol octopamine. Animals were both observed and videotaped for 15 min and observed overnight following injection. Behavioral quantification was performed by analyzing the video using the open-source software ZebraZoom (https://github.com/oliviermirat/ZebraZoom; Mirat et al. 2013) to track the center-of-mass of the snails. The quality of the tracking was assessed visually to confirm the correct position. The data were visualized using ggplot in R using 1 frame per second to calculate the distance traveled. For each animal, the distance traveled was measured every 5 min giving a total of three measurements. Each of the 5 min measurements was compared with the same time point in the control. Two sided t-test was done using base R to calculate significance.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge assistance from the Ministry of Environment, Climate Change, Disaster Management, and Meteorology (Solomon Islands) and Solomon Islands National University for collection and sequencing of Conus tmetus. The collection of this species was made under the permit number RP/2017/003 granted to the University of Utah by the Solomon Islands government. The authors also acknowledge the assistance from the Marine Science Institute of the University of the Philippines in the collection of the remaining Conus specimens. The collection was made possible under permit number 0252-23 granted by the Philippine Department of Agriculture-Bureau of Fisheries and Aquatic Resources. The HEK293-hNET cell line was a generous gift from the Blakely Lab in Vanderbilt University. The authors acknowledge Bob Schackman from the University of Utah DNA/Peptide Core facility for sequencing χ-AuId and William Low from the Salk Institute for the MALDI mass analysis. The support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged. Samuel Abalde offered assistance for conducting the BEAST analysis.
Contributor Information
Samuel Espino, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
Maren Watkins, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
Rodolfo Probst, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA; Science Research Initiative, College of Science, University of Utah, Salt Lake City, UT 84112, USA.
Thomas Lund Koch, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA; Department of Biochemistry, University of Utah, Salt Lake City, UT 84112, USA; Department of Biomolecular Sciences, University of Copenhagen, Copenhagen N 2200, Denmark.
Kevin Chase, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
Julita Imperial, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
Samuel D Robinson, Institute for Molecular Bioscience, The University of Queensland, St Lucia, QLD 4072, Australia.
Paula Flórez Salcedo, Department of Neurobiology, University of Utah, Salt Lake City, UT 84112, USA.
Dylan Taylor, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
Joanna Gajewiak, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
Mark Yandell, Eccles Institute of Human Genetics, University of Utah, Salt Lake City, UT 84112, USA; Utah Center for Genetic Discovery, University of Utah, Salt Lake City, UT 84112, USA.
Helena Safavi-Hemami, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA; Department of Biochemistry, University of Utah, Salt Lake City, UT 84112, USA; Department of Biomolecular Sciences, University of Copenhagen, Copenhagen N 2200, Denmark.
Baldomero M Olivera, School of Biological Sciences, University of Utah, Salt Lake City, UT 84112, USA.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Funding
This work was supported in part by the National Institute of General Medical Science (NIGMS) grant to B.M.O. and M.Y. (R01GM122869), a grant from NIGMS awarded to B.M.O. and H.S.-H. (R01GM144719), and a Villum Foundation Young Investigator Grant 19063 to H.S.-H.
Data Availability
Accession numbers for all the 2-Loop Tau conopeptides are available in Supplementary Table 2. Sequences and relevant files used for Phylogenetic analysis, Divergence timing analysis, Clustering analysis and Phylogenetic and selection analysis of T-superfamily conotoxins are available in figshare and can be accessed through the links provided in the method section.
References
- Abalde S, Dutertre S, Zardoya R. A combined transcriptomics and proteomics approach reveals the differences in the predatory and defensive venoms of the molluscivorous cone snail Cylinder ammiralis (Caenogastropoda: Conidae). Toxins (Basel). 2021:13(9):642. 10.3390/toxins13090642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abalde S, Tenorio MJ, Uribe JE, Zardoya R. Conidae phylogenomics and evolution. Zool Scr. 2019:48(2):194–214. 10.1111/zsc.12329. [DOI] [Google Scholar]
- Adamo SA. Norepinephrine and octopamine: linking stress and immune function across phyla. Invert Surv J. 2008:5:12–19. 10.1093/icb/icu005. [DOI] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990:215(3):403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aonuma H, Mezheritskiy M, Boldyshev B, Totani Y, Vorontsov D, Zakharov I, Ito E, Dyakonova V. The role of serotonin in the influence of intense locomotion on the behavior under uncertainty in the mollusk Lymnaea stagnalis. Front Physiol. 2020:11:221. 10.3389/fphys.2020.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji RA, Ohtake A, Sato K, Gopalakrishnakone P, Kini RM, Seow KT, Bay B-H. λ-conotoxins, a new family of conotoxins with unique disulfide pattern and protein folding. Isolation and characterization from the venom of Conus marmoreus. J Biol Chem. 2000:275(50):39516–39522. 10.1074/jbc.M006354200. [DOI] [PubMed] [Google Scholar]
- Barghi N, Concepcion GP, Olivera BM, Lluisma AO. Comparison of the venom peptides and their expression in closely related Conus species: insights into adaptive post-speciation evolution of Conus exogenomes. Genome Biol Evol. 2015:7(6):1797–1814. 10.1093/gbe/evv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely RD, Berson HE, Fremeau RT Jr, Caron MG, Peek MM, Prince HK, Bradley CC. Cloning and expression of a functional serotonin transporter from rat brain. Nature. 1991:354(6348):66–70. 10.1038/354066a0. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchene S, Fourment M, Gavryushkina A, Heled J, Jones G, Kuhnert D, De Maio N, et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2019:15(4):e1006650. 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw CD. Miocene climates. In: Alderton D, Elias S, editors. Encyclopedia of geology. Cambridge, Massachusetts, USA: Academic Press; 2021. p. 486–496. [Google Scholar]
- Broer S, Gether U. The solute carrier 6 family of transporters. Br J Pharmacol. 2012:167(2):256–278. 10.1111/j.1476-5381.2012.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust A, Palant E, Croker DE, Colless B, Drinkwater R, Patterson B, Schroeder CI, Wilson D, Nielsen CK, Smith MT, et al. χ-Conopeptide pharmacophore development: toward a novel class of norepinephrine transporter inhibitor (Xen2174) for pain. J Med Chem. 2009:52(22):6991–7002. 10.1021/jm9003413. [DOI] [PubMed] [Google Scholar]
- Chase K, Watkins M, Safavi-Hemami H, Olivera BM. Integrating venom peptide libraries into a phylogenetic and broader biological framework. Front Mol Biosci. 2022:9:784419. 10.3389/fmolb.2022.784419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AG, Norberg T, Griffin D, Hoeger C, Akhtar M, Schmidt K, Low W, Dykert J, Richelson E, Navarro V, et al. Contulakin-G, an O-glycosylated invertebrate neurotensin. J Biol Chem. 1999:274(20):13752–13759. 10.1074/jbc.274.20.13752. [DOI] [PubMed] [Google Scholar]
- Duda TF Jr, Kohn AJ, Palumbi SR. Origins of diverse feeding ecologies within Conus, a genus of venomous marine gastropods. Biol J Linn Soc. 2001:73(4):391–409. 10.1111/j.1095-8312.2001.tb01369.x. [DOI] [Google Scholar]
- Duda TF Jr, Palumbi SR. Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc Natl Acad Sci U S A. 1999:96(12):6820–6823. 10.1073/pnas.96.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda TF Jr, Palumbi SR. Gene expression and feeding ecology: evolution of piscivory in the venomous gastropod genus Conus. Proc Biol Sci. 2004:271(1544):1165–1174. 10.1098/rspb.2004.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Jin AH, Kaas Q, Jones A, Alewood PF, Lewis RJ. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol Cell Proteomics. 2013:12(2):312–329. 10.1074/mcp.M112.021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre S, Jin AH, Vetter I, Hamilton B, Sunagar K, Lavergne V, Dutertre V, Fry BG, Antunes A, Venter DJ, et al. Evolution of separate predation- and defence-evoked venoms in carnivorous cone snails. Nat Commun. 2014:5(1):3521. 10.1038/ncomms4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espino SS, Robinson SD, Safavi-Hemami H, Gajewiak J, Yang W, Olivera BM, Liu Q. Conopeptides promote itch through human itch receptor hMgprX1. Toxicon. 2018:154:28–34. 10.1016/j.toxicon.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu DJ, Watkins M, Dia-Monje V, Cartier GE, Cruz LJ, Olivera BM. Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon. 2001:39(12):1899–1916. 10.1016/S0041-0101(01)00175-1. [DOI] [PubMed] [Google Scholar]
- Espiritu MJ, Taylor JK, Sugai CK, Thapa P, Loening NM, Gusman E, Baoanan ZG, Baumann MH, Bingham J-P. Characterization of the native disulfide isomers of the novel chi-conotoxin PnID: implications for further increasing conotoxin diversity. Mar Drugs. 2023:21(2):61. 10.3390/md21020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A, DeFelice LJ, Duke B-J, Moore KR, Blakely RD. Sodium-dependent norepinephrine-induced currents in norepinephrine-transporter-transfected HEK-293 cells blocked by cocaine and antidepressants. J Exp Biol. 1995:198(Pt 10):2197–2212. 10.1242/jeb.198.10.2197. [DOI] [PubMed] [Google Scholar]
- Gillette R. Evolution and function in serotonergic systems. Integr Comp Biol. 2006:46(6):838–846. 10.1093/icb/icl024. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011:29(7):644–652. 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010:59(3):307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Gupta K, Kumar M, Balaram P. Disulfide bond assignments by mass spectrometry of native natural peptides: cysteine pairing in disulfide bonded conotoxin. Anal Chem. 2010:82(19):8313–8319. 10.1021/ac101867e. [DOI] [PubMed] [Google Scholar]
- Hackney CM, Florez Salcedo P, Mueller E, Koch TL, Kjelgaard LD, Watkins M, Zachariassen LG, Tuelung PS, McArthur JR, Adams DJ, et al. A previously unrecognized superfamily of macro-conotoxins includes an inhibitor of the sensory neuron calcium channel Cav2.3. PLoS Biol. 2023:21(8):e3002217. 10.1371/journal.pbio.3002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018:35(2):518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junier T, Zdobnov EM. The newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010:26(13):1669–1670. 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas Q, Westermann J-C, Halai R, Wang CK, Craik DJ. ConoServer, a database for conopeptide sequences and structures. Bioinformatics. 2008:24(3):445–446. 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017:14(6):587–589. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013:30(4):772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch TL, Ramiro IBL, Florez Salcedo P, Engholm E, Jensen KJ, Chase K, Olivera BM, Bjorn-Yoshimoto WE, Safavi-Hemami H. Reconstructing the origins of the somatostatin and allatostatin-C signaling systems using the accelerated evolution of biodiverse cone snail toxins. Mol Biol Evol. 2022:39(4):msac075. 10.1093/molbev/msac075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch TL, Robinson SD, Salcedo PF, Chase K, Biggs J, Fedosov AE, Yandell M, Olivera BM, Safavi-Hemami H. Prey shifts drive venom evolution in cone snails. Mol Biol Evol. 2024:41(8):msae120. 10.1093/molbev/msae120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn AJ. The ecology of Conus in Hawaii. Ecol Monogr. 1959:29(1):47–90. 10.2307/1948541. [DOI] [Google Scholar]
- Kowalewski M. Drill holes produced by the predatory gastropod Nucella lamellosa (Muricidae): palaeobiological and ecological implications. J Molluscan Studies. 2004:70(4):359–370. 10.1093/mollus/70.4.359. [DOI] [Google Scholar]
- Lee Y, Park J-K. Complete mitochondrial genome of Conus lischkeanus Weinkauff, 1875 (Neogastropoda, Conidae) and phylogenetic implications of the evolutionary diversification of dietary types of Conus species. Zookeys. 2022:1088:173–185. 10.3897/zookeys.1088.78990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JL, Lukowiak K. The behavior of Aplysia californica cooper (Gastropoda; Opisthobranchia): I. Ethogram. Behavior. 1986:98(1-4):320–360. 10.1163/156853986X01035. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Dutertre S, Vetter I, Christie MJ. Conus venom peptide pharmacology. Pharmacol Rev. 2012:64(2):259–298. 10.1124/pr.111.005322. [DOI] [PubMed] [Google Scholar]
- Li Q, Watkins M, Robinson SD, Safavi-Hemami H, Yandell M. Discovery of novel conotoxin candidates using machine learning. Toxins (Basel). 2018:10(12):503. 10.3390/toxins10120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey S, Carew TJ. Locomotion in Aplysia: triggering by serotonin and modulation by bag cell extract. J Neurosci. 1983:3(7):1469–1477. 10.1523/JNEUROSCI.03-07-01469.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinesco S, Wickremasinghe N, Kolkman KE, Carew TJ. Serotonergic modulation in aplysia. II. Cellular and behavioral consequences of increased serotonergic tone. J Neurophysiol. 2004:92:2487–2496. 10.1152/jn.00210.2004. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Corpuz GO, Layer RT, Garrett JE, Wagstaff JD, Bulaj G, Vyazovkina A, Yoshikami D, Cruz LJ, Olivera BM. Isolation and characterization of a novel Conus peptide with apparent antinociceptive activity. J Biol Chem. 2000:275(42):32391–32397. 10.1074/jbc.M003619200. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020:37(5):1530–1534. 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirat O, Sternberg JR, Severi KE, Wyart C. ZebraZoom: an automated program for high-throughput behavioral analysis and categorization. Front Neural Circuits. 2013:7:107. 10.3389/fncir.2013.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen CK, Lewis RJ, Alewood D, Drinkwater R, Palant E, Patterson M, Yaksh TL, McCumber D, Smith MT. Anti-allodynic efficacy of the χ-conopeptide, Xen2174, in rats with neuropathic pain. Pain. 2005:118(1):112–124. 10.1016/j.pain.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Olivera BM. E.E. Just lecture, 1996. Conus venom peptides, receptor and ion channel targets, and drug design: 50 million years of neuropharmacology. Mol Biol Cell. 1997:8(11):2101–2109. 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera BM. Conus venom peptides: reflections from the biology of clades and species. Annu Rev Ecol Syst. 2002:33(1):25–47. 10.1146/annurev.ecolsys.33.010802.150424. [DOI] [Google Scholar]
- Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006:281(42):31173–31177. 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Cruz LJ. Conotoxins, in retrospect. Toxicon. 2001:39(1):7–14. 10.1016/S0041-0101(00)00157-4. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Gray WR, Zeikus R, McIntosh JM, Varga J, Rivier J, de Santos V, Cruz LJ. Peptide neurotoxins from fish-hunting cone snails. Science. 1985:230(4732):1338–1343. 10.1126/science.4071055. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Seger J, Horvath MP, Fedosov AE. Prey-capture strategies of fish-hunting cone snails: behavior, neurobiology and evolution. Brain Behav Evol. 2015:86(1):58–74. 10.1159/000438449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczkowski FA, Sharpe IA, Dutertre S, Lewis RJ. χ-Conotoxin and tricyclic antidepressant interactions at the norepinephrine transporter define a new transporter model. J Biol Chem. 2007:282(24):17837–17844. 10.1074/jbc.M610813200. [DOI] [PubMed] [Google Scholar]
- Pahari A, Mondal S, Bardhan S, Sarkar D, Saha S, Buragohain D. Subaerial naticid gastropod drilling predation by Natica tigrina on the intertidal molluscan community of Chandipur, eastern coast of India. Palaeogeogr Palaeoclimatol Palaeoecol. 2016:451:110–123. 10.1016/j.palaeo.2016.03.020. [DOI] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004:20(2):289–290. 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pardos-Blas JR, Irisarri I, Abalde S, Tenorio MJ, Zardoya R. Conotoxin diversity in the venom gland transcriptome of the magician's cone, pionoconus magus. Mar Drugs. 2019:17(10):553. 10.3390/md17100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardos-Blas JR, Tenorio MJ, Galindo JCG, Zardoya R. Comparative venomics of the cryptic cone snail species Virroconus ebraeus and Virroconus judaeus. Mar Drugs. 2022:20(2):149. 10.3390/md20020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuong MA, Mahardika GN. Targeted sequencing of venom genes from cone snail genomes improves understanding of conotoxin molecular evolution. Mol Biol Evol. 2018:35(5):1210–1224. 10.1093/molbev/msy034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puillandre N, Bouchet P, Duda TF Jr, Kauferstein S, Kohn AJ, Olivera BM, Watkins M, Meyer C. Molecular phylogeny and evolution of the cone snails (Gastropoda, Conoidea). Mol Phylogenet Evol. 2014:78:290–303. 10.1016/j.ympev.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst Biol. 2018:67(5):901–904. 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratibou Z, Inguimbert N, Dutertre S. Predatory and defensive strategies in cone snails. Toxins (Basel). 2024:16(2):94. 10.3390/toxins16020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SD, Norton RS. Conotoxin gene superfamilies. Mar Drugs. 2014:12(12):6058–6101. 10.3390/md12126058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockel D, Korn W, Kohn AJ. Manual of the living Conidae. Wiesbaden, Germany: Verlag Christa Hemmen; 1995. [Google Scholar]
- Rogalski A, Himaya SWA, Lewis RJ. Coordinated adaptations define the ontogenetic shift from worm- to fish-hunting in a venomous cone snail. Nat Commun. 2023:14(1):3287. 10.1038/s41467-023-38924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi-Hemami H, Gajewiak J, Karanth S, Robinson SD, Ueberheide B, Douglass AD, Schlegel A, Imperial JS, Watkins M, Bandyopadhyay PK, et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc Natl Acad Sci U S A. 2015:112(6):1743–1748. 10.1073/pnas.1423857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol Biol Evol. 2002:19(1):101–109. 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011:27(6):863–864. 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe IA, Gehrmann J, Loughnan ML, Thomas L, Adams DA, Atkins A, Palant E, Craik DJ, Adams DJ, Alewood PF. et al. et al. Two new classes of conopeptides inhibit the α1-adrenoceptor and noradrenaline transporter. Nat Neurosci. 2001:4(9):902–907. 10.1038/nn0901-902. [DOI] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 2015:32(5):1342–1353. 10.1093/molbev/msv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006:34(Web Server issue):W609–W612. 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007:56(4):564–577. 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004:84(1):41–68. 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Terlau H, Shon K-J, Grilley M, Stocker M, Stuhmer W, Olivera BM. Strategy for rapid immobilization of prey by a fish-hunting marine snail. Nature. 1996:381(6578):148–151. 10.1038/381148a0. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium . UniProt: a hub for protein information. Nucleic Acids Res. 2015:43(D1):D204–D212. 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe JE, Puillandre N, Zardoya R. Beyond Conus: phylogenetic relationships of Conidae based on complete mitochondrial genomes. Mol Phylogenet Evol. 2017:107:142–151. 10.1016/j.ympev.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Walker CS, Steel D, Jacobsen RB, Lirazan MB, Cruz LJ, Hooper D, Shetty R, DelaCruz RC, Nielsen JS, Zhou LM, et al. The T-superfamily of conotoxins. J Biol Chem. 1999:274(43):30664–30671. 10.1074/jbc.274.43.30664. [DOI] [PubMed] [Google Scholar]
- Williams ST, Duda TF Jr. Did tectonic activity stimulate oligo-miocene speciation in the Indo-West Pacific? Evolution. 2008:62(7):1618–1634. 10.1111/j.1558-5646.2008.00399.x. [DOI] [PubMed] [Google Scholar]
- WoRMSEditorialBoard [Internet] . 2023. [accessed 2023 Aug 15]. https://www.marinespecies.org.
- Wu Y, Wang L, Zhou M, You Y, Zhu X, Qiang Y, Qin M, Luo S, Ren Z, Xu A. Molecular evolution and diversity of Conus peptide toxins, as revealed by gene structure and intron sequence analyses. PLoS One. 2013:8(12):e82495. 10.1371/journal.pone.0082495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018:19(Suppl 6):153. 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers for all the 2-Loop Tau conopeptides are available in Supplementary Table 2. Sequences and relevant files used for Phylogenetic analysis, Divergence timing analysis, Clustering analysis and Phylogenetic and selection analysis of T-superfamily conotoxins are available in figshare and can be accessed through the links provided in the method section.