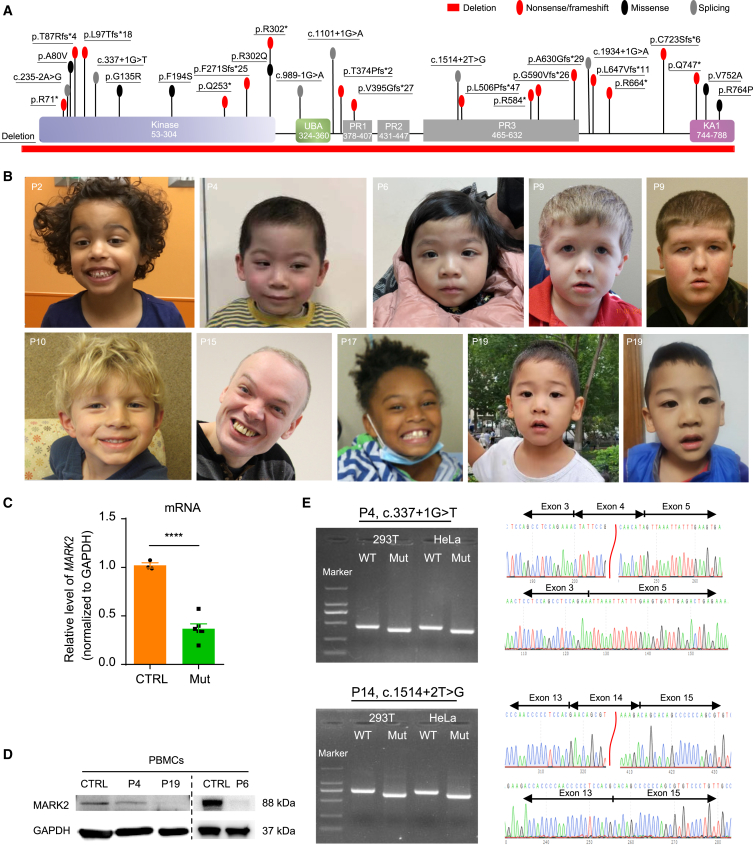
Figure 1.
Overview of MARK2 variants and associated phenotypes
(A) Locations of variants in functional domains of MARK2 predicted by UniProt are shown. p.Arg302∗ and p.Gln747∗ are recurrent variants that occurred in 2 individuals. Kinase, protein kinase domain; UBA, ubiquitin associated; PR, polar residue; KA1, kinase-associated domain 1.
(B) Facial photos of 8 affected individuals. Ages at the time of photograph are P2, 5 years and 4 months; P4, 4 years and 2 months; P6, 3 years and 1 month; P9 left, 4 years; P9 right, 11 years; P10, 8 years; P15, 46 years; P17, 7 years and 7 months; P19 left, 4 years; P19 right, 4 years and 7 months.
(C and D) The mRNA expression (C) and protein accumulation (D) of MARK2 in the PBMCs from 3 affected individuals (Mut: P4, P6, P19). Both mRNA and protein levels were normalized to GAPDH levels and compared with those of age-matched healthy children (CTRL). The data of at least 3 independent experiments were analyzed by Student’s t test, ∗p < 0.05.
(E) Minigene assays of 2 splicing variants (P4: c.337+1G>T; P14: c.1514+2T>G). RT‒PCR products from HEK 293T and HeLa cells transfected with either the wild-type (WT) or mutant (Mut) pcDNA3.1 vector were separated by electrophoresis (left). M, DNA marker; the sizes of the bands are 2,000, 1,000, 750, 500, 250, and 100 bp. Sequencing of the RT-PCR products (right) show c.337+1G>T variant results in a 49-bp deletion, while the c.1514+2T>G variant results in a 98-bp deletion.