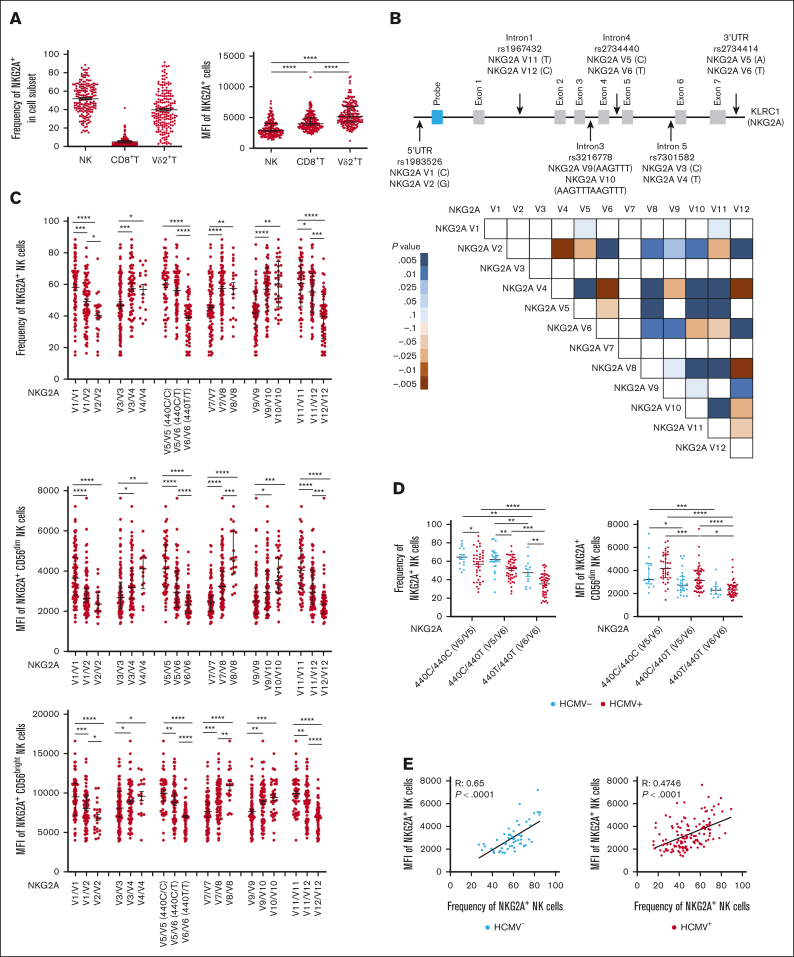
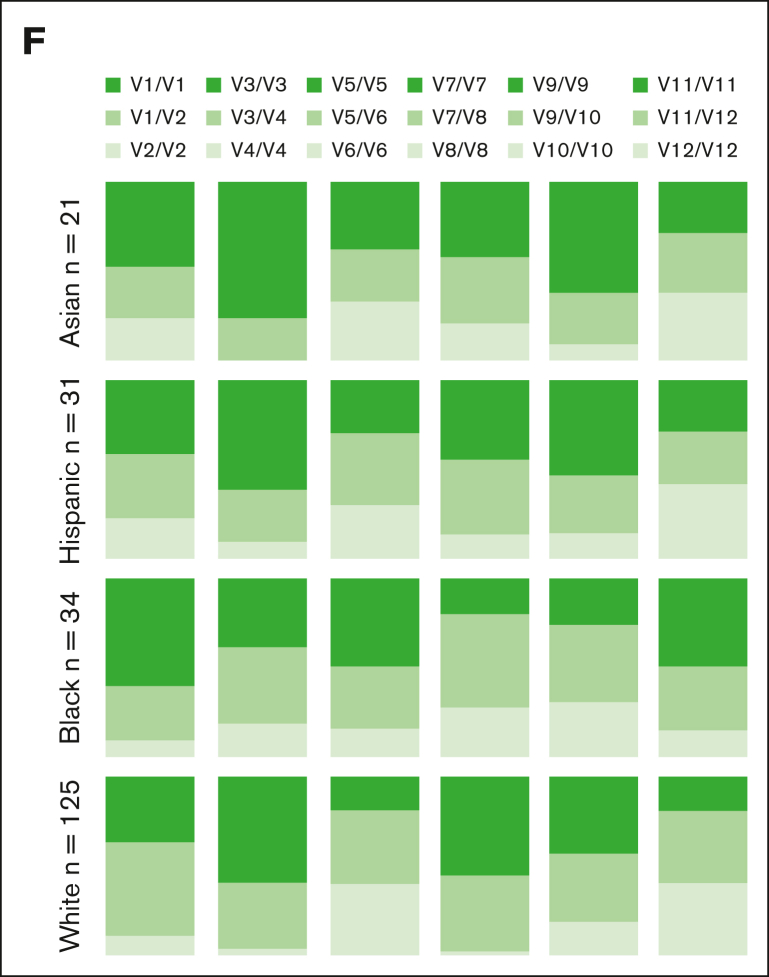
Figure 1.
Common NKG2A noncoding SNPs are in positive LD and associate with NKG2A expression independent of HCMV serostatus. (A) NKG2A frequency and MFI in NK cells, CD8+ T cells, and Vδ2+ T cells from phenotyping of 204 healthy blood donors. (B) Common noncoding KLRC1 SNPs and LD analysis from genotyping of 204 donors. Positive LD shaded blue; negative LD shaded red. (C) NKG2A+ NK cell frequency and MFI stratified by NKG2A variant. (D) NKG2A+ NK cell frequency and MFI stratified by HCMV serostatus and SNP rs2734440 (440C refers to SNP rs2734440 C and 440T refers to rs2734440 T). (E) Correlation between NKG2A+ NK cell frequency and MFI in HCMV– and HCMV+ individuals. (F) Distribution of rs2734440 alleles among different ethnic groups. For panels C-D, t tests were performed, and the mean ± standard error of the mean (SEM) was presented to analyze NKG2A frequencies. For panels A,C-D, Mann-Whitney tests were performed, and median ± interquartile range (IQR) is presented to analyze NKG2A MFI. Correlations were assessed using the Pearson correlation coefficient for panel E. LD was assessed using the χ2 test for panel B. The symbols represent individual samples. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.