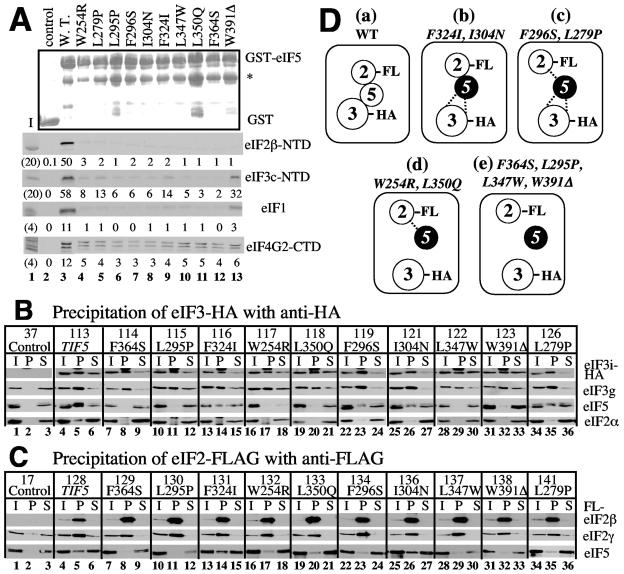
FIG. 3.
Effect of the eIF5-CTD Ts− mutation on factor interactions in vivo and in vitro. (A) Effect of the Ts− mutations on binding of eIF5 to MFC partners in vitro. Lanes 2 to 13, ∼4 μg of GST-eIF5 or its indicated mutant derivatives were incubated with 35S-eIF2β-NTD (second panel), -eIF3c-NTD (third panel), -eIF1 (fourth panel), or -eIF4G-CTD (fifth panel) in 100 μl of binding buffer. The protein complex was isolated using glutathione Sepharose and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (40). GST-eIF5 and its mutants were expressed and purified from Escherichia coli BL21(DE3) carrying pGEX-TIF5 or the corresponding pGEX-tif5 derivatives (Table 2). 35S-labeled proteins were expressed in the TnT system (Promega) from pT7-SUI3ΔS, pHis-NIP1-N, pT7-SUI1, and pT7-TIF4632ΔS. Relative amounts of 35S-labeled proteins bound to the resin were determined by phosphorimaging analyses of sodium dodecyl sulfate-polyacrylamide gels and are shown below each panel. Top panel, Coomassie staining pattern; bottom four panels, autoradiography. Asterisk at the top panel indicates the position of a GST-eIF5 cleavage product resulting from bacterial protease activity. Lanes 1, 20% (second and third panels) or 4% (fourth and fifth panels) input amount. (B and C) Two hundred micrograms of whole-cell extract prepared from KAY yeast strains with indicated tif5 mutations, grown in yeast extract-peptone-dextrose medium, was used for immunoprecipitation with anti-HA (panel B) or anti-FLAG (panel C) affinity resin, as described previously (40). The entire pellet fractions (P) were analyzed, along with 10% input (I) and supernatant (S) fractions, by immunoblotting using polyclonal anti-eIF5 (24), anti-eIF3g (38), anti-eIF2α (13), and anti-eIF2γ (18) antibodies as indicated to the right. Monoclonal anti-HA (BabCO) and anti-FLAG (Sigma) antibodies were used to detect eIF3i-HA and FLAG-eIF2β, in panels B and C, respectively. Control, KAY37 (TIF5 TIF34) in panel B and KAY17 (TIF5 SUI3) in panel C; TIF5+ strains encoded untagged eIF3i and eIF2β, respectively (see reference 3 for detailed genotypes). Numbers above the TIF5/tif5 allele designations refer to the relevant KAY yeast strains. In the bottom row of panel B, the bands smaller and larger than eIF2α in lanes 8, 11, 14, and 17 represent cross-reactive species detected by anti-HA antibodies and do not represent eIF2α. (D) Model of MFC assembly in the strains tested. Circles indicate individual eIFs. Filled circles indicate the mutant eIF5. HA and FL refer to HA and FLAG epitope tags introduced to eIF2 and eIF5, respectively. Direct contact indicates strong interactions, equivalent in level to that seen with the respective wild-type strain. The dotted line indicates weak interactions, i.e., ∼30 to 50% of the level seen with the wild type. No line between the factors indicates that the interaction was not detected by the experiments in panels B and C.