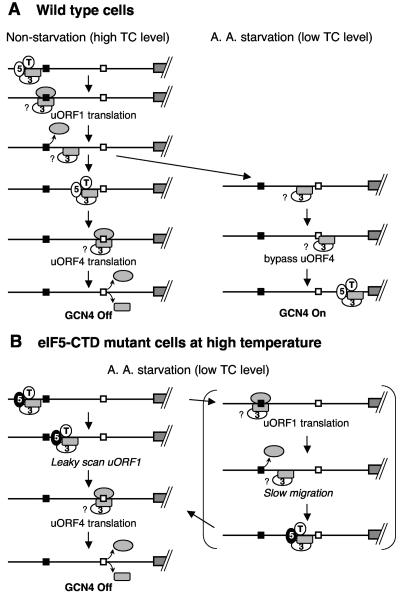
FIG. 7.
Model for GCN4 translational control in wild-type and eIF5-CTD mutant cells. Lines indicate the GCN4 mRNA leader, and the grey boxes to the right (followed by diagonal hashed lines) represent the GCN4 coding region. Two regulatory uORFs (uORFs 1 and 4) have been shown to be necessary and sufficient for regulation of GCN4 expression, and are depicted as filled and open squares, respectively. The figure illustrates the ribosome movement on the leader region with the focus on its association with eIF3 (numeral 3), eIF5 (numeral 5), and eIF2 TC (T). The 40S and 60S subunits are drawn as grey rectangles with rounded corners and as grey ovals, respectively. (A) Control GCN4 translation in wild-type cells. Under nonstarvation conditions (left column), the preinitiation complex scans for and translates uORF1. Evidence suggests that eIF3 is associated with the ribosome at this stage (see the text for details). Question marks indicate uncertainties relative to these associations. Following uORF1 translation, a population of 40S subunits remains associated with the mRNA and resumes scanning after reacquiring TC and other eIFs (third and fourth panels). Subsequent translation of uORF4 dissociates the ribosome, shutting off GCN4 translation. Under amino acid (A. A.) starvation conditions (right column), Gcn2p kinase is activated and phosphorylates eIF2. This phosphorylation renders eIF2 into a competitive inhibitor of GDP/GTP exchange activity (catalyzed by eIF2B; not shown), thereby reducing the level of eIF2/GTP and hence TC levels. uORF4 is bypassed due to low TC levels. TC can be reacquired during scanning of the uORF4-to-GCN4 interval, resulting in translation of GCN4. (B) In eIF5-CTD mutants, impaired eIF5-CTD function at a higher temperature confers a Gcn− phenotype. The figure illustrates possible mechanisms for this observation. Mutant eIF5 (5) is depicted as filled ovals. (Left column) The preinitiation complex with the mutant eIF5 scans past the uORF1 start codon (leaky scanning) and instead initiates at the uORF4 start codon. Translation of uORF4 leads to efficient ribosome dissociation, shutting off GCN4 translation (17). This mechanism differs from that proposed to explain the Gcn−phenotype in prt1-1 mutants. In the latter case (right column), the 40S subunit tethered by uORF1 translation migrates very slowly and receives TC prior to the uORF4 start codon. The slowed migration may compensate for levels of TC reduced by eIF2 phosphorylation (34).