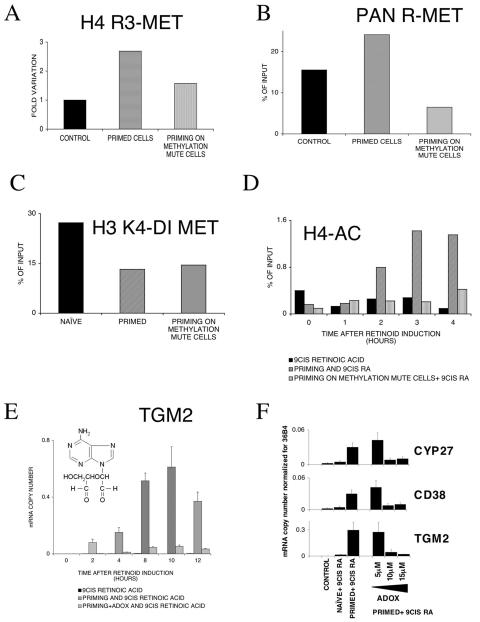
FIG. 6.
Epigenetic changes on TGM2 promoter and mRNA expression upon priming in methylation mute cells (methyltransferases were blocked with 10 μM ADOX for 16 h as described in Materials and Methods). Values are the means of three parallel QPCR measurements ± standard deviations, and results were confirmed from at least three independent biological samples. All chromatin immunoprecipitation values are the means of three independent QPCR measurements of a representative experiment. Chromatin immunoprecipitation results were confirmed in at least three independent chromatin preps. (A) H4 arginine 3 methylation levels determined by chromatin immunoprecipitation on the HR1 enhancer element. (B) Arginine methylation levels determined by chromatin immunoprecipitation on the HR1 enhancer element with an anti-Pan methyl arginine antibody. (C) H3 lysine 4 methylation studied by chromatin immunoprecipitation on the core promoter. (D) Chromatin immunoprecipitation analysis of H4 acetylation levels in naive, primed, and primed methylation mute cells on the enhancer element HR1. (E) TGM2 mRNA levels normalized to 36B4 copy numbers in naive, primed, and primed methylation mute cells (the structure of ADOX is shown in the inset). (F) TGM2, CD38, and CYP27 transcript levels after blocking methylation with ADOX in naive and primed cells. ADOX was used in increasing concentrations as shown. The mRNA copy numbers were normalized to 36B4 transcript levels. Values are the means of three parallel QPCR measurements ± standard deviations, and results were confirmed from at least three independent biological samples.