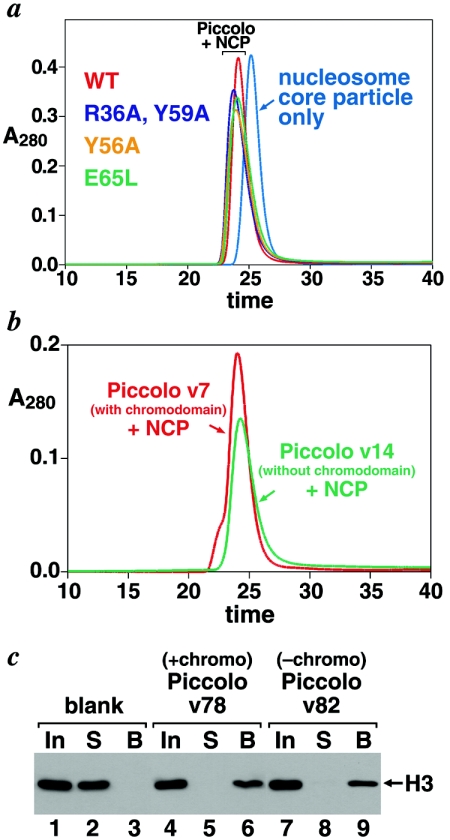
FIG. 6.
Esa1 chromodomain is not necessary for Piccolo to bind nucleosomes. (a) Gel filtration chromatograms of recombinant nucleosome core particles alone (cyan) and nucleosome core particles incubated with Piccolo v7 (red), Piccolo v72 (blue), Piccolo v69 (yellow), and Piccolo v74 (green). All Piccolo samples form stable complexes with nucleosomes, as shown by the appearance of a larger peak, distinct from the nucleosome peak and from the Piccolo-only peak at approximately 27 min (data not shown). SDS-PAGE gels of the peaks confirm the assignment of the gel filtration peaks. For example, fractions for the Piccolo/nucleosome peaks show all three Piccolo subunits and all four nucleosome histone subunits (data not shown). (b) Gel filtration chromatograms of recombinant nucleosome core particles (NCP) incubated with Piccolo v7 (red), which contains full-length Esa1, or Piccolo v14 (green), which lacks the Esa1 chromodomain. Both samples form the Piccolo/nucleosome complex, establishing that the chromodomain is not required for stable binding of Piccolo to nucleosomes. (c) Piccolo/nucleosome pulldown experiment confirms that the Esa1 chromodomain is not necessary for binding to nucleosomes. Blank (lanes 1 to 3), Piccolo v78 (lanes 4 to 6), or v82 (lanes 7 to 9) was immobilized on Strep-Tactin beads via their Strep-tagged Yng2 subunits and incubated with nucleosome core particles, and unbound and bound fractions were analyzed by Western blots to detect the histone H3 component of nucleosomes. Input samples are shown in lanes 1, 4, and 7; unbound (supernatant) fractions in lanes 2, 5, and 8; and bound fractions in lanes 3, 6, and 9.