Abstract
The type I receptor-like protein tyrosine kinase MuSK is essential for the neuromuscular junction formation. MuSK expression is tightly regulated during development, but the underlying mechanisms were unclear. Here we identified a novel mechanism by which MuSK expression may be regulated. A cyclic AMP response element (CRE)-like element in the 5′-flanking region of the MuSK gene binds to CREB1 (CRE-binding protein 1). Mutation of this element increases the MuSK promoter activity, suggesting a role for CREB1 in attenuation of MuSK expression. Interestingly, CREB mutants unable to bind to DNA also inhibit MuSK promoter activity, suggesting a CRE-independent inhibitory mechanism. In agreement, CREB1 could inhibit a mutant MuSK transgene reporter whose CRE site was mutated. We provide evidence that CREB interacts directly with MyoD, a myogenic factor essential for MuSK expression in muscle cells. Suppression of CREB expression by small interfering RNA increases MuSK promoter activity. These results demonstrate an important role for CREB1 in the regulation of MuSK expression.
MuSK, a type I receptor-like protein tyrosine kinase, is a central organizer in the formation of the neuromuscular junction (NMJ) (40). MuSK mutant mice do not form the NMJ (10, 27, 53), and muscle cells derived from MuSK mutant mice do not form acetylcholine receptor (AChR) clusters in response to agrin (16). Expression of MuSK is under tight spatial and temporal control. Like the AChR, the mRNA of MuSK is enriched at the NMJ (39, 42, 50). During development, MuSK mRNA is detectable in rat myotomes, but not dermatomes, at embryonic day 11 (E11), when myotomes form (50). The expression level continues to increase and peaks around E16 to E18, and after that declines slowly to a barely detectable level in adult muscle (23, 28).
Molecular mechanisms underlying MuSK expression is complex. Like many muscle-specific genes, expression of MuSK requires E box elements which stimulate the promoter activity when binding to myogenic factors such as MyoD (24). Several factors appear to enhance MuSK expression. For example, MuSK promoter activity or mRNA was increased in muscle cells stimulated with neuregulin (23, 25), a factor that increases AChR expression via the mitogen-activated protein kinase pathways (1, 45, 46, 49). Downstream of mitogen-activated protein kinases is the Ets transcription factor GABP, which appears to be necessary and sufficient for synaptic expression (15, 41, 43). Transgenic mice expressing a dominant-negative Ets mutant demonstrate alterations in NMJ postsynaptic morphology and decreased expression of genes containing an Ets-binding site; however, the expression of MuSK and Rapsyn was unchanged (11). These data may suggest that MuSK expression is regulated by an Ets-independent mechanism. We reported recently that the MuSK promoter could be up-regulated by Wnt, a factor implicated in the NMJ formation in Drosophila melanogaster (24). Exactly how Wnt stimulates MuSK expression has yet to be determined since Wnt stimulates multiple signaling pathways, including the small GTPase Rac. Rac activation appears to be required for MuSK expression (25).
In this study, we investigated the regulation of MuSK expression by CREB (cyclic AMP [cAMP] response element [CRE]-binding protein). A CRE-like element in the 5′-flanking region of the MuSK gene binds to CREB1 to inhibit the MuSK promoter activity. Further characterization revealed that CREB mutants unable to bind to DNA were also able to inhibit MuSK promoter activity, suggesting a CRE-independent inhibitory mechanism. We show that CREB could interact directly with the myogenic factor MyoD. Suppression of CREB expression by small interfering RNA (siRNA) increased MuSK promoter activity. These results demonstrate an important role for CREB1 in the regulation of MuSK expression.
MATERIALS AND METHODS
Plasmid constructs and antibodies.
Antibodies against CREB1 (sc-186), p300 (sc-585), and MyoD (sc-760) were bought from Santa Cruz (Santa Cruz, CA). Anti-myosin heavy chain (anti-MHC) antibody (MF-20) was from the Developmental Studies Hybridoma Bank. Forskolin (final concentration, 10 μM) and 8-bromo-cAMP (final concentration, 1 mM) were purchased from Calbiochem and dissolved in dimethyl sulfoxide. CREB1 and deletion mutants were amplified by PCR with Pfu Turbo with rat CREB1 as the template (Stratagene, La Jolla, CA) and subcloned between BamHI and EcoRI in pKH3 in frame downstream from the three-hemagglutinin (HA) epitope. MyoD and deletion mutants were amplified using mouse MyoD as the template with Pfu Turbo and subcloned between EcoRI and XbaI in pCS2+ in frame downstream of the six-Myc epitope. M715(mt)-Luc, whose CRE-like sequence was changed from TTA TGT CA (−648 to −641) to GGC GTG AC, was produced using the Quick Change site-directed mutagenesis kit. Two artificial reporters of M215, which has only one E box (24), were generated to add a consensus CRE sequence (underlined), GGG AGA GAT TGC CTG ACG TCA GAG AGC TAG GGT AC, or a mutant CRE sequence, GGG AGA GAT TGC CGA CCA TAG GAG AGC TAG GGT AC, into the cloning site between SmaI and KpnI of the M215 reporter.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed according to the protocol provided by Upstate Biotechnology (Lake Placid, NY). C2C12 myotubes were washed twice with phosphate-buffered saline (PBS) and then incubated in 1% formaldehyde in PBS at room temperature for 10 min. After washing twice with PBS, cells were scraped in PBS containing protease inhibitors including 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 2 μg/ml aprotinin. Cells were collected by a brief centrifugation at 2,000 rpm at 4°C and were resuspended in sodium dodecyl sulfate (SDS) lysis buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.0), 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, 1 μg/ml leupeptin, and 2 μg/ml aprotinin. Cells were sonicated by Sonic Dismembrator model 100 (Fisher, Suwanee, GA), strength 3, for 15 s three times, to shear chromosome DNA to 500 to 1,000 bp in length, which was monitored by gel electrophoresis. After centrifugation at 13,000 at 4°C, the sonicated mixture was diluted 10-fold with ChIP dilution buffer containing 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), and 167 mM NaCl. The mixture was precleaned once with salmon sperm DNA-protein A-agarose beads at 4°C for 1 h. The resulting supernatant was incubated with indicated antibodies at 4°C overnight, when salmon sperm DNA-protein A-agarose beads (50 μl) was added and the mixture was incubated for another hour at 4°C. Beads were washed sequentially with a low-salt wash buffer containing 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), and 150 mM NaCl; a high-salt wash buffer containing 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), and 500 mM NaCl; an LiCl wash buffer containing 0.25 M LiCl, 1% NP-40, 1% deoxycholic acid, 1 mM EDTA, and 10 mM Tris-HCl (pH 8.1); and TE buffer. Beads were then incubated twice with 250 μl of the elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min at room temperature to elute bound DNA. Combined eluates were added to 20 μl of 5 M NaCl to reverse cross-links by heating at 65°C for 4 h. After incubation with 10 μl of 0.5 M EDTA, 20 μl of 1 M Tris-HCl (pH 6.5), and 2 μl of 10 mg/ml proteinase K (1 h at 45°C), DNA was recovered by phenol-chloroform extraction and ethanol precipitation.
PCRs were conducted with the eluted DNA as the template and the following primers. For the MuSK 5′-flanking region (694 bp), the sense primer was −758 5′CTT GGA AAC AAT TCA CTG CCA GCA GG and the antisense primer was −64 5′CAA TAG AGT TGT CCG CTA AGG ACT GTT TG (24). For the AChRα subunit 5′-flanking region (237 bp), the sense primer was 5′GAC AAG CCT CTG ACT CAT GAT CTA TGT and the antisense primer was 5′GCT GCC GGT CCT ACT CCA CCC TGG CT (47). For the AChRɛ C-terminal coding region (310 bp), the sense primer was 5′GTG TTT GAG GGT CAG AGG CA and the antisense primer was 5′TGC AGG CTC ATG GTT GGA TG.
EMSA.
The electrophoretic mobility shift assay (EMSA) was modified from Choi et al. (8). His-CREB1 was purified and incubated in 20 μl (final volume) of the DNA-binding buffer (10 mM HEPES [pH 7.9], 50 mM KCl, 0.5 mM dithiothreitol, 5% glycerol) supplemented with 1 μg of poly(dI-dC) and 1 ng of 32P-labeled double-stranded oligonucleotide probe (∼10,000 cpm). After incubation at 30°C for 15 min, the reaction mixture was run onto a 6% acrylamide gel in 0.5× Tris-borate-EDTA buffer at 4°C. In some reaction mixtures, unlabeled double-stranded oligonucleotides or anti-CREB antibody was included. The CRE probe sequence was 5′AGA CAA GTA GAT GCC AGC GAG TCC CCT TAT GTC ACA GCA GAG AGG ATT GC (the CRE-like element is underlined). EMSA probes in the −215 5′-flanking region were 5′TGT TCC TCC AGC CCT CCA CTT GCA TTA CTG GCC TGA GTG AGT TCT CCC CAG CTG GCC ACG TCC (M215A), 5′ACG TCC ATC ATT ACA GCC AGG TTT GAG GAT TCC TGA TTC TAC TTA GAG CCG GTG GCG CTG CAG (M215B), 5′CGC TGC AGA CAC AAA CAG TCC TTA GCG GAC AGC TCT ATT GTA ACA ACC ATG CTT TAA AAT GTA (M215C), and 5′AAA ATG TAA ACC CGG GAG CGT GTT TTT TTG GTT TTT TTT TTT TTC CTC ACG TTG TCC AG (M215D).
Cell culture, transfection, luciferase assay, and immunoprecipitation.
C2C12 cells were maintained as described previously (45). Myoblasts (80% confluency) were transiently cotransfected with a MuSK promoter-luciferase reporter and pRL-CMV (as a control for transfection efficiency and sample handling) in a ratio of 20:1 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. In some experiments, cotransfection included mutant CREB1 and/or MyoD (in a ratio of 100:20:1, mutant CREB1 and/or MyoD to reporter to pRL-CMV). Twenty-four hours after transfection, myoblasts were switched to the differentiation medium as described previously (45) and luciferase assays were performed after myotubes were fully developed using the dual-luciferase kit (Promega, Madison, WI) as previously described (24). The firefly luciferase activity was normalized to Renilla luciferase activity. Immunoprecipitation and immunoblotting were performed as previously described (29). Unless otherwise indicated, all experiments were repeated at least three times.
Pulldown assay.
Radiolabeled CREB1 proteins were generated by in vitro translations using the SP6 coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine. 35S-labeled CREB1 proteins were incubated with glutathione S-transferase (GST)-MyoD immobilized on beads in the binding buffer (25 mM HEPES [pH 7.5], 1 mM dithiothreitol, 0.5% Triton X-100, 150 mM NaCl, protease inhibitors) for 2 h at 4°C on a rotator. After washing with PBS-0.1% Tween 20, bound CREB1 proteins were subjected to SDS-polyacrylamide gel electrophoresis and Western blotting.
Knockdown of CREB1 expression by siRNA.
CREB1-siRNA and control oligonucleotides were purchased from Ambion (catalog no. 16704). C2C12 myoblasts were transfected with respective oligonucleotides together with report genes and/or MyoD with Lipofectamine 2000. Luciferase activities were assayed as described above. The effect of CREB1-siRNA on CREB1 expression was determined by cotransfecting COS-7 cells with the respective oligonucleotides and HA-tagged CREB1. Expression of CREB1 was monitored by immunoblotting.
RNase protection assays.
A 184-nucleotide probe spanning exon1 and exon2 of the MuSK gene was subcloned in pGEM-T (Promega). The plasmid was linearized with NcoI. Radiolabeled antisense RNA probe was produced with SP6 RNA polymerase and [α-32P]UTP using a MAXIscript kit (Ambion) following the manufacturer's instructions. 32P-labeled MuSK and actin probes were mixed with 20 μg total RNA of C2C12 myotubes in RNase protection assay II hybridization buffer (80% deionized formamide, 100 mM sodium citrate [pH 6.4], 300 mM sodium acetate, 1 mM EDTA) at 42°C overnight. After digestion with RNase, protected fragments were fractionated on 6% polyacrylamide sequencing gel.
RESULTS
An inhibitory CRE-like element in the MuSK promoter.
In analyzing the MuSK promoter, we found that the MuSK promoter contains a CRE-like element between −648 and −641 (Fig. 1A) (24). This element contains nucleotides that are necessary for CREB binding (34). CRE elements have been implicated in negative regulation of the MyoD transactivation activity (36). We were interested in testing whether the CRE-like element in the MuSK promoter has a similar function. First we determined whether CREB proteins bind to the CRE-like element using ChIP assays. As shown in Fig. 1B, MuSK DNA fragments encompassing the CRE region were detectable in the precipitates brought down by anti-CREB1 antibodies. No band was revealed if no antibody was added to the reaction mixture. Moreover, immunoprecipitation with the anti-CREB1 antibody did not precipitate DNA fragments of the 5′-flanking region of the AChR α subunit gene, which does not contain the CRE cis element (47), or the AChR ɛ subunit coding region (Fig. 1B). Antibodies against p300 also precipitated the MuSK promoter (Fig. 1B). p300 is a histone acetyltransferase that is known to interact with CREB1 and MyoD and play a role in regulating gene expression (37, 54). It is conceivable that p300 association with the MuSK promoter may be indirect, i.e., via MyoD and/or CREB1 (32).
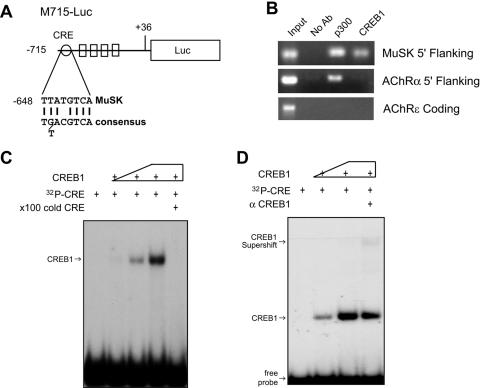
FIG. 1.
Identification of a CRE-like element in the MuSK promoter. (A) Schematic diagram of M715-Luc. The CRE-like element, located between −648 and −641, has the consensus sequence of the CRE element (24). (B) Interaction of the CRE-like element with CREB1 in ChIP assays. Chromatin preparations of C2C12 myotubes were incubated without (No Ab) or with antibodies against CREB1 or p300. Immunopurified DNA was used as the template for PCR amplification using primers against the indicated products. (C) Binding of the CRE-like element to purified CREB1. A 50-bp DNA fragment flanking the CRE-like element of the MuSK promoter was labeled with [γ-32P]ATP by PNK. The labeled probe was incubated with various amounts of purified His-CREB1 without or with excess wild-type unlabeled probes. The reaction mixture was resolved on a nondenaturing polyacrylamide gel. (D) Supershift of the CRE probe-CREB1 complex in the presence of anti-CREB1 antibodies.
To determine whether the CRE cis element binds directly to CREB1, EMSA was performed using a radioactive DNA probe encompassing the CRE-like element. Figure 1C shows the binding of purified recombinant CREB1 to the probe in a dose-dependent manner. The binding was suppressed when an excess of unlabeled probe was present. Moreover, the addition of anti-CREB1 antibody caused a slower migration of the complex (or supershift), suggesting the bound protein is CREB1 (Fig. 1D).
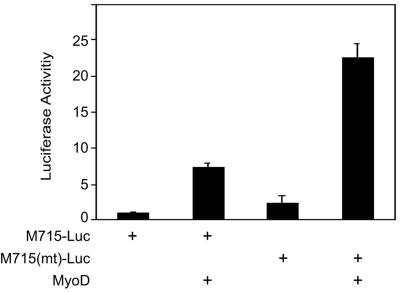
To study the function of the CRE-like element in regulation of MuSK promoter activity, we mutated it by replacing thymidines (T), adenosines (A), cytidines (C), and guanosines (G) with G, C, A, and T, respectively. The mutant probe was unable to compete with the wild type in binding to nuclear proteins (data not shown). The basal promoter activity of M715-Luc was increased in C2C12 myotubes when the element was mutated [M715(mt)-Luc], suggesting a negative role for the element. We showed earlier that MyoD increases the promoter activity of the MuSK gene when coexpressed in C2C12 myotubes (Fig. 2) (24). The MyoD-mediated enhancement was further elevated in M715(mt)-Luc, further supporting the notion that the element may be inhibitory. Next we explored the effects of CREB expression on MuSK promoter activity to determine if the transcription factor functions through binding to the CRE-like element. As shown in Fig. 3A, CREB expression inhibited MyoD-induced promoter activity in M715-Luc and M715(mt)-Luc. These results corroborate to suggest that MuSK expression may be inhibited by the CREB/CRE pathway.
FIG. 2.
Mutation of the CRE-like element enhances promoter activity. The CRE-like element in M715(mt)-Luc was mutated to 5′G GCG TGA C. C2C12 myoblasts were transfected with pRL-CMV and M715-Luc or M715(mt)-Luc. Firefly and Renilla luciferase activities were assayed in C2C12 myotubes. Shown is the relative luciferase activity (firefly/Renilla, mean ± the standard error of the mean) from a representative experiment done in duplicate and repeated three times with similar results.
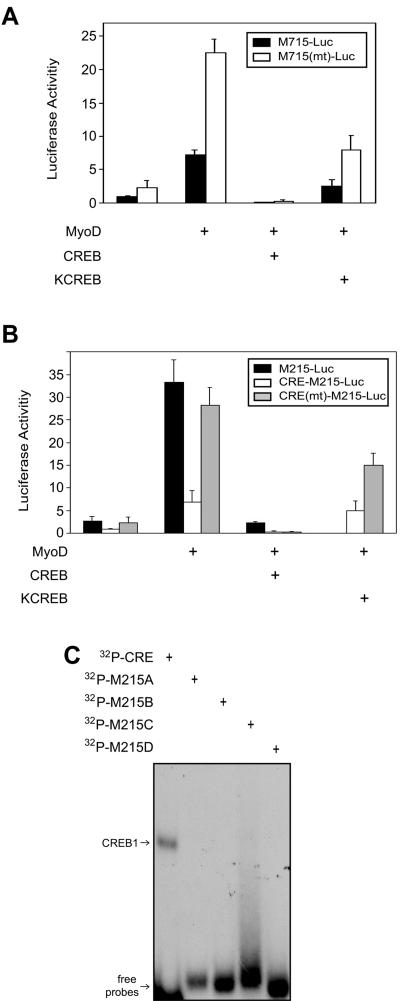
FIG. 3.
Inhibitory effects of KCREB and CREB on MyoD transactivation of the MuSK promoter. (A) Down-regulation by KCREB and CREB of MyoD transactivation. M715-Luc or M715(mt)-Luc was cotransfected with pRL-CMV and pCMV-CREB or pCMV-KCREB. Luciferase activities were assayed as in Fig. 2. (B) Identification of theE box 1 element as the site by which CREB or KCREB inhibits MuSK promoter activity. M215-Luc contains the 215-bp 5′-flanking region and the luciferase gene. CRE-M215-Luc contains the CRE element (5′GGG AGA GAT TGC CTG ACG TCA GAG AGC TAG GGT AC), the 215-bp 5′-flanking region, and the luciferase gene. In CRE(mt)-M215-Luc, the CRE element was mutated to 5′GGG AGA GAT TGC CGA CCA TAG GAG AGC TAG GGT AC (mutated portion underlined). Luciferase activities were assayed as in panel A. Shown is relative luciferase activity (firefly/Renilla, mean ± the standard error of the mean) from a representative experiment done in duplicate and repeated three times with similar results. (C) No CREB1-binding activity was detected in the −215 5′-flanking region. DNA fragments encompassing the −215 5′-flanking region (M215A-D; see Materials and Methods for sequence information) or the CRE cis element were labeled as in Fig. 1 and incubated with purified CREB1. The reaction was resolved on nondenaturing polyacrylamide gel.
CRE-independent inhibition of MuSK promoter activity.
If CREB1 inhibits MuSK promoter activity by binding to the CRE element, mutation of the DNA-binding domain in CREB1 should prevent it from executing the effect. We used KCREB, a mutant that is unable to bind to the CRE element and functions as a dominant repressor of the wild-type factor (51). As shown in Fig. 3A, CREB1's inhibitory activity was decreased, supporting the notion that CREB1 binds to the CRE element to inhibit MuSK expression. However, the MuSK promoter activity enhanced by MyoD in cells expressing KCREB was lower than the control. This result was unexpected, which could suggest that DNA binding may not be entirely necessary for CREB1's inhibitory effect. To test this hypothesis, we studied the effect of CREB on M715(mt)-Luc, where the CRE-like element was mutated. CREB remained able to inhibit M715(mt)-Luc, demonstrating a CRE-independent inhibitory mechanism (Fig. 3A). M715-Luc contains several E box elements, the CRE element, a MEF2-like element, and a C/EBP-like element (24).
To eliminate the involvement of other elements, we created an artificial reporter (CRE-M215-Luc) containing a consensus CRE sequence (TG ACG TCA), the 215 5′-flanking region of the MuSK gene, and the luc gene (see Materials and Methods for details). The 215-bp 5′-flanking region contains the E box that is necessary for muscle-specific expression of MuSK but does not contain any other identifiable cis element (24). The addition of the CRE sequence suppressed the basal, as well MyoD-upregulated, promoter activity (Fig. 2B). This inhibition was reversed when the CRE element was mutated [CRE(mt)-M215-Luc], suggesting the suppressive nature of the CRE element. As with M715-Luc, CREB and KCREB inhibited MyoD-mediated expression of CRE-M215-Luc (Fig. 3B). Mutation of the CRE element did not prevent this inhibitory effect, suggesting a DNA binding-independent mechanism. To eliminate the possibility that CREB1 may still bind to DNA within the −215 5′-flanking region, we generated a series of four DNA probes to cover the entire region. As shown in Fig. 3C, none bound recombinant CREB1 except the probe containing the CRE site at −648. These results agreed with the data from studies of M715-Luc that CREB may regulate MuSK expression without binding to the CRE element.
Coimmunoprecipitation between CREB and MyoD.
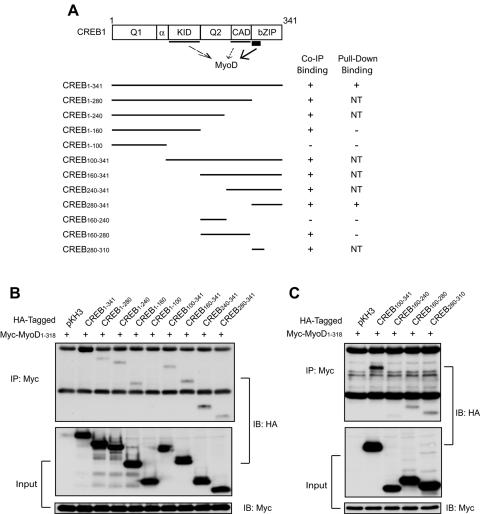
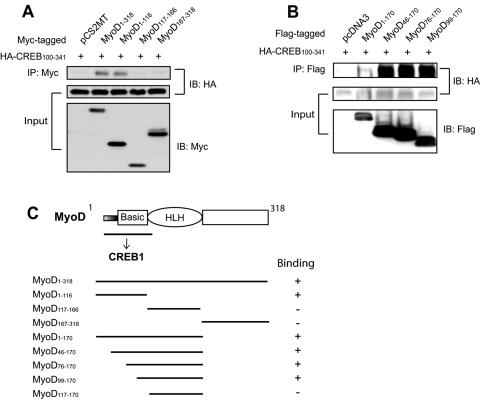
Since the 215-bp 5′-flanking region contains only E box 1, we reasoned that CREB regulates MuSK expression probably via interacting with MyoD. Thus, we tested whether the two proteins form a complex in cells. Myc-tagged full-length MyoD and HA-tagged CREB constructs were coexpressed in COS-7 cells and characterized for association by immunoprecipitation. As shown in Fig. 4B (lane 2), full-length CREB was detectable in precipitates by anti-HA antibodies, suggesting that the two proteins may form a complex in cells. CREB has five domains, Q1 (glutamine-rich transcription activator domain 1), KID (kinase-inducible domain), Q2 (glutamine-rich transcription activator domain 2), CAD (constitutive activation domain), and bZIP (basic leucine zipper) (44). MyoD associated with various C-terminal deletion mutants except CREB1-100 (lane 6), indicating that the N-terminal 100 amino acid residues are disposable (Fig. 4A and B). Studies with N-terminal deletion mutants confirmed that the binding activity resides in the C-terminal region. However, CREB160-240, which encompasses the Q2 domain, was not detectable in MyoD precipitates (Fig. 4C). In summary, the KID, CAD, and bZIP domains of CREB associated with MyoD in immunoprecipitation assays (Fig. 4A).
FIG. 4.
MyoD coimmunoprecipitated with multiple domains of CREB1. (A) Schematic diagram of CREB1 constructs and their binding activities. Putative binding domains are underlined; thin lines indicate indirect binding, whereas the thick line indicates direct binding (see Fig. 6). Co-IP binding, coimmunoprecipitation data from panels B and C; pull-down binding, data from Fig. 6; NT, not determined. (B and C) Coimmunoprecipitation between MyoD and CREB1 and deletion mutants. COS-7 cells were transfected with Myc-tagged MyoD and HA-tagged CREB1 or its deletion mutants. Twenty-four hours after transfection, cells were lysed and subjected to immunoprecipitation with anti-Myc antibody. CREB proteins in the immunocomplex were revealed by immunoblotting with anti-HA antibody. The bottom two panels show inputs of HA-CREB1 constructs and Myc-MyoD, respectively. IP, immunoprecipitation; IB, immunoblotting.
We performed reciprocal precipitation experiments to identify binding domains in MyoD. Because CREB association with MyoD requires the C-terminal region, CREB100-341 was coexpressed with various MyoD constructs in COS-7 cells (Fig. 5). CREB1 appeared to associate with the N-terminal region of MyoD but not the helix-loop-helix (HLH) domain or the C-terminal region (Fig. 5B). To further map the region, a series of deletions in the basic domain were tested for binding to CREB1. MyoD1-170 associated with CREB1, whereas MyoD117-170 did not, suggesting that the binding activity is located in the first 117 amino acid residues. Thus, sequences in the N-terminal region, in addition to the basic domain, may be involved in binding to CREB1. These results are in line with an earlier report that MyoD may form a complex with CREB to regulate the activity of the retinoblastoma protein promoter (31).
FIG. 5.
MyoD interacts with CREB1 via the basic domain. (A and B) Coimmunoprecipitation between CREB1 and MyoD and its deletion mutants. COS-7 cells were transfected with plasmids expressing the indicated proteins, i.e., HA-tagged CREB100-341, which contains the C-terminal region, and Myc-tagged MyoD or its deletion mutants. Immunoprecipitation was performed as in Fig. 4. CREB proteins in the immunocomplex were revealed by immunoblotting with anti-HA antibody. The bottom two panels show inputs of HA-CREB1 and Myc-MyoD constructs, respectively. (C) Schematic diagram of MyoD constructs and their CREB1 binding activities. IP, immunoprecipitation; IB, immunoblotting.
Direct interaction between MyoD and CREB1.
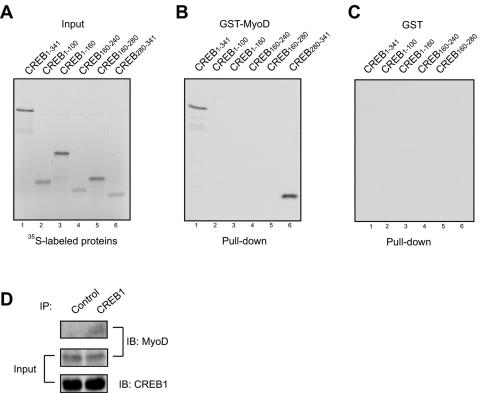
To determine whether MyoD interacts directly with CREB1 and not via a third protein, recombinant CREB1 or its fragments were generated by in vitro translation in the presence of [35S]methionine (Fig. 6A). 35S-labeled CREB was incubated with recombinant MyoD fused with GST that was immobilized on agarose beads. GST alone did not precipitate with CREB1 (lane 1, Fig. 6C). In contrast, GST-MyoD pulled down 35S-labeled full-length CREB (lane 1, Fig. 6B), suggesting that the two proteins interact directly. To map the domain in CREB that binds to MyoD, various CREB1 recombinant proteins were generated and incubated with GST-MyoD immobilized on beads. In accord with results from immunoprecipitation studies, CREB1-100 (Q1 domain) and CREB160-243 (Q2 domain) did not bind MyoD (Fig. 6B). In contrast, GST-MyoD was able to pull down only CREB280-341, the region that contains the bZIP domain, suggesting that this domain is sufficient to bind to MyoD. Interestingly, although CREB1-160 coimmunoprecipitated with MyoD (Fig. 4), it did not appear to interact with MyoD in pulldown assays, suggesting that the binding of CREB1-160 to MyoD may be indirect, possibly via p300. CREB1 was detectable in precipitates with anti-MyoD antibodies, suggesting that the two proteins interact in vivo (Fig. 6D).
FIG. 6.
Direction interaction between MyoD and CREB1. (A) CREB1 and the indicated deletion proteins were generated by in vitro translation in the presence of [35S]methionine using the TNT SP6 coupled reticulocyte lysate system (Promega). 35S-labeled proteins were revealed by autoradiography. (B) Direct interaction of MyoD with the bZIP domain in CREB1. 35S-labeled CREB1 constructs were incubated with recombinant GST-MyoD protein (full length) immobilized on agarose beads. After washing, bead-bound proteins were resolved by SDS-polyacrylamide gel electrophoresis and autoradiography. (C) GST-beads did not interact with 35S-labeled CREB1 constructs. (D) Interaction between endogenous MyoD and CREB1 in muscle cells. Lysates of C2C12 cells were incubated with anti-CREB1 antibody or an irrelevant antibody (control), and resulting complexes were probed with anti-MyoD antibody. IP, immunoprecipitation; IB, immunoblotting.
Increased MuSK promoter activity in muscle cells expressing CREB1-siRNA.
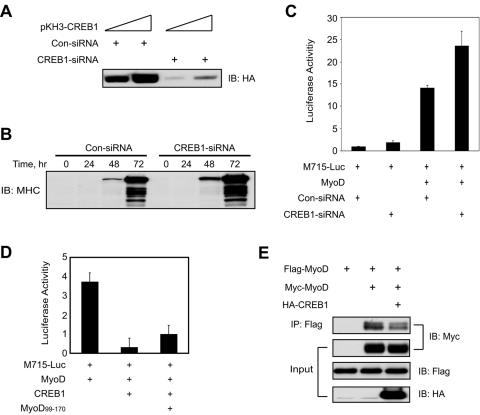
To determine the regulatory role of CREB1 in regulating MuSK expression in vivo, we suppressed expression of endogenous CREB1 by using an oligonucleotide-based siRNA strategy (Fig. 7A) (21). Suppression of CREB1 expression appeared to increase the expression of MHC, a marker of muscle differentiation, suggesting that CREB1 may be a negative regulator of muscle differentiation (Fig. 7B). To study the effect of CREB1 suppression on MuSK promoter activity, CREB1-siRNA was transfected with M715-Luc and MyoD into C2C12 myoblasts. At 24 h after transfection, myoblasts were induced to form myotubes. Luciferase activity was assayed 24 h after myotube formation. As shown in Fig. 7C, MyoD enhancement of the MuSK promoter activity was increased by CREB1-siRNA in comparison with control siRNA. These results suggest an inhibitory role for endogenous CREB1 in regulation of MuSK expression in muscle cells.
FIG. 7.
Regulation of MuSK promoter activity by endogenous CREB1. (A) CREB1-siRNA inhibited CREB1 expression. COS-7 cells were transfected with pKH3-CREB1 together with CREB1-siRNA or control siRNA (Con-siRNA) oligonucleotides. Expression of transfected CREB1 was revealed by immunoblotting with anti-HA antibody. (B) Effects of suppression of CREB1 expression on muscle differentiation. Transfected C2C12 myotubes were collected at different times after being switched to the differentiation medium and lysed. Lysates were probed for expression of MHC. (C) Increased MuSK promoter activity in C2C12 myotubes cotransfected with CREB1-siRNA. M715-Luc was cotransfected with pRL-CMV and pCMV-MyoD with CREB1-siRNA or Con-siRNA. Luciferase activities were assayed as in Fig. 2. Shown is relative luciferase activity (firefly/Renilla, mean ± the standard error of the mean) from a representative experiment done in duplicate and repeated three times with similar results. (D) Expression of MyoD99-170 reversed CREB1 inhibition of MyoD transactivation activity. M715-Luc was cotransfected with pRL-CMV and pCMV-MyoD with MyoD99-170. Luciferase activities were assayed as in Fig. 2. Shown is relative luciferase activity (firefly/Renilla, mean ± the standard error of the mean) from a representative experiment done in duplicate and repeated three times with similar results. (E). Expression of CREB1 inhibited dimerization of MyoD. Flag- or Myc-tagged MyoD was coexpressed without or with HA-CREB1 in COS-7 cells. Lysates were incubated with anti-Flag antibody and resulting lysates probed with anti-Myc antibody. IP, immunoprecipitation; IB, immunoblotting.
We determined whether the inhibition of CREB-MyoD interaction prevents the repressive effect of CREB1. MyoD-binding activity for CREB1 appeared to reside in a minimal region around amino acids 99 to 116 (Fig. 5). Expression of MyoD99-170, a deletion mutant containing the minimal interaction region that is able to interact with CREB1, could reverse CREB1 inhibition of MyoD transactivation activity (Fig. 7D), presumably by preventing endogenous CREB1 from interacting with MyoD. To explore inhibitory mechanisms of CREB1, we looked at the effect of CREB1 on MyoD dimerization because CREB binds to a region close to that necessary for dimerization. Flag-MyoD and Myc-MyoD were transfected into COS-7 cells. Lysates were incubated with anti-Flag antibody, and resulting immunocomplexes were probed with anti-Myc antibody. Myc-MyoD was detectable in precipitates with anti-Flag antibody, suggesting that MyoD may form a homodimer (Fig. 7E). The amount of Myc-MyoD in the complex was decreased in cells expressing CREB1, suggesting that the latter may inhibit the formation of MyoD homodimers. These results could suggest that CREB1 attenuates MyoD transactivation activity by interfering with its dimerization.
Effects of forskolin and bromo-cAMP on MuSK expression.
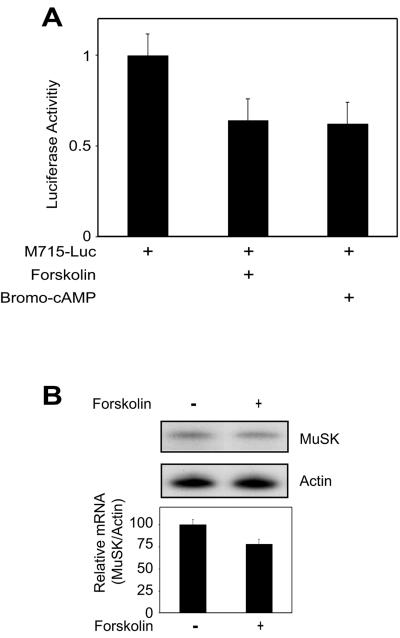
The activation of the cAMP pathway is implicated in regulation of the expression of synapse-specific genes at the NMJ (13). An increase in intracellular cAMP or activation of the cAMP-dependent protein kinase increases the level of AChR and acetylcholinesterase mRNA (7, 9, 14). In light of these observations, we looked at effects of forskolin on MuSK expression. Forskolin is a stimulator of adenylate cyclase (12). Treatment with forskolin inhibited MuSK-Luc expression and decreased the level of MuSK mRNA in muscle cells (Fig. 8). Similar effects were observed with bromo-cAMP, a protein kinase A (PKA) activator (Fig. 8). These results suggest that activation of the cAMP pathway, presumably by factors released from motoneurons (such as CGRP) (13), may regulate MuSK expression.
FIG. 8.
Effects of forskolin and bromo-cAMP on expression of the M715-Luc transgene and MuSK mRNA. (A) C2C12 myotubes transfected with M715-Luc and pRL-CMV were treated with forskolin, bromo-cAMP, or vehicle (−) for 24 h. Luciferase activities were assayed as in Fig. 2. Shown is relative luciferase activity (firefly/Renilla, mean ± the standard error of the mean) from a representative experiment done in duplicate and repeated three times with similar results. (B) C2C12 myotubes were treated with forskolin for 24 h and lysed for RNA purification. RNase protection assay was performed as described in Materials and Methods. Shown are representative blots (top) and histograms summarizing three independent experiments (mean ± the standard error of the mean).
DISCUSSION
Our previous studies presented evidence that MuSK expression is controlled by myogenic factors that bind to E box elements in the 5′-flanking region (24). Here we identify a CRE-like element in the 5′-flanking region of the MuSK gene that binds to CREB1 and inhibits the MuSK promoter activity. We also show that CREB1 inhibits MuSK expression by a CRE-independent mechanism. CREB1 inhibited a mutant MuSK transgene reporter whose CRE site was mutated, and CREB mutants unable to interact with DNA also inhibit MuSK promoter activity. Finally, suppression of CREB expression by siRNA increased MuSK promoter activity. These results demonstrate an important role for CREB1 in the regulation of MuSK expression.
CREB1 belongs to a multigene family with several isoforms that may function as transcriptional activators or repressors (19). While a large number of genes are up-regulated by CREB proteins, recent studies suggest that these factors could suppress gene expression. For example, transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB (55). The insulin gene has two CRE-like elements in the 5′-flanking region (12). Forskolin, presumably via cAMP, suppresses insulin gene transcription and transcriptional activity of insulin gene reporter in the INS-1 beta-cell line (12). Our study presents evidence that CREB1 could inhibit MuSK gene transcription by two different mechanisms: one dependent on binding to a CRE element and the other in a manner independent of DNA-binding activity. The first notion is supported by three lines of evidence. First, CREB1 binds to the CRE-like element in ChIP and mobility shift assays (Fig. 1). Second, expression of CREB1 suppresses the MuSK promoter activity (Fig. 3). Third, mutation of the CRE-like element increases the promoter activity (Fig. 2).
KCREB, a mutant that does not bind DNA, remains able to inhibit the MuSK promoter activity (Fig. 3). Moreover, CREB1 expression was able to inhibit a MuSK transgene that does not have the CRE-like element (Fig. 3). Further studies indicate that CREB inhibition requires the E box element, which is known to interact with MyoD (24). These results suggest that CREB1 may regulate MuSK expression via myogenic factors. Indeed, we provided evidence that CREB1 could interact with MyoD in vitro and in vivo. Knockdown in CREB1 expression increases MuSK promoter activity in muscle cells. Taken together, these results suggest a second mechanism by which CREB1 inhibits MuSK expression, which is likely mediated by direct interaction between CREB1 and MyoD. It is interesting that CREB could function as the DNA-recognizing factor of the MyoD-responsive element in the RB promoter, whereas the DNA-binding function of MyoD is dispensable (31).
The mechanism by which CREB1 inhibits MyoD transactivation activity remains unclear. MyoD activity is regulated by numerous interacting proteins such as p300, retinoic acid receptor, and MEF2. These proteins promote transactivation by interaction with the basic domain or the N terminus of MRF transcription factors (33, 35, 54). Serum response factor, Sp1, muscle LIM protein, and the product of the retinoblastoma gene (pRb) increase MyoD activity by interacting with the HLH domain (2, 4, 17, 18). The C terminus of MyoD could bind to TFIID, which may facilitate the recruitment of TFIIB in the preinitiation complex (20). In addition to positive regulators, MyoD forms a complex with proteins that decrease the transactivation activity. They include Id, Twist, MyoR, Mist1, c-Jun, Fos, and JunB, which interact with the HLH domain (38). On the other hand, the nuclear receptor corepressor interacts with the N terminus in MyoD to inhibit transactivation activity (3). CREB1 interacts not with the HLH domain or the C terminus but the basic region of MyoD. I-mf proteins are myogenic suppressors that could inhibit MyoD binding to DNA (6). Because the region in MyoD for CREB1 interaction binds DNA, one plausible hypothesis is that CREB1 may interfere with the DNA-binding activity of MyoD in a manner similar to I-mf (inhibitor of MyoD family) proteins. We characterized MyoD binding to E box elements in gel shift assays in the presence or absence of increasing concentrations of CREB1 and failed to observe an effect of CREB1 on MyoD-DNA interaction (data not shown). This result may suggest that CREB1 may function by a different mechanism. Since the CREB-binding site in MyoD is close to the region necessary for MyoD dimerization (30), we reasoned that CREB could attenuate MyoD transactivation activity by interfering with its dimerization. This notion is supported by data in Fig. 7E, providing a potential mechanism of CREB1 action.
Transcription of genes encoding synapse-specific proteins is suppressed in extrasynaptic nuclei. It is believed that calcium influx stimulated by electric activity activates various kinases including PKA, PKC, and CaMK (22, 42, 48, 52). Phosphorylation of MyoD decreases its transactivation activity (5, 22, 26, 48). Phosphorylation and/or activation of CREB1 by calcium influx may stimulate its recruitment to the MuSK transcription machinery. It is worth pointing out that both the CREB protein level and the level of phosphorylation of the CREB protein at Ser-133 rapidly increase at the onset of muscle differentiation and that both remain high throughout the myogenic process (31). This expression pattern, together with results of this paper, provides an explanation for the low expression of MuSK in adult muscles.
The activation of the cAMP pathway is also implicated in regulation of the expression of synapse-specific genes at the NMJ (13). Augmentation of cAMP levels or activation of the cAMP-dependent protein kinase promotes expression of the AChR and acetylcholinesterase (7, 9, 14). The identification of the CRE-like element in the MuSK promoter and the inhibition by CREB1 may provide a novel mechanism to regulate MuSK expression. Considering that CREB1 may function as a downstream component in the cAMP pathway, it is conceivable that MuSK may be regulated by factors affecting levels of cAMP. Indeed, muscle cells treated with forskolin or bromo-cAMP appeared to show less MuSK promoter activity or mRNA (Fig. 8). These results suggest that activation of the cAMP pathway, presumably by factors released from motoneurons (such as CGRP) (13), may regulate MuSK expression. The negative regulatory mechanism might prevent MuSK from being overexpressed and spilled over into the extrasynaptic area.
Acknowledgments
This work is support in part by grants from the National Institutes of Health (L.M. and W.C.X.), the Muscular Dystrophy Association (L.M.), and the Philip Morris External Research Program (L.M.).
We are grateful to Karl Tsim and Roland P. S. Kwok for CREB1 constructs.
REFERENCES
- 1.Altiok, N., S. Altiok, and J. P. Changeux. 1997. Heregulin-stimulated acetylcholine receptor gene expression in muscle: requirement for MAP kinase and evidence for a parallel inhibitory pathway independent of electrical activity. EMBO J. 16:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber, S., G. Halder, and P. Caroni. 1994. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell 79:221-231. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, P., M. Downes, P. Lau, J. Harris, S. L. Chen, Y. Hamamori, V. Sartorelli, and G. E. Muscat. 1999. The nuclear receptor corepressor N-CoR regulates differentiation: N-CoR directly interacts with MyoD. Mol. Endocrinol. 13:1155-1168. [DOI] [PubMed] [Google Scholar]
- 4.Biesiada, E., Y. Hamamori, L. Kedes, and V. Sartorelli. 1999. Myogenic basic helix-loop-helix proteins and Sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac α-actin promoter. Mol. Cell. Biol. 19:2577-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blagden, C. S., L. Fromm, and S. J. Burden. 2004. Accelerated response of the myogenin gene to denervation in mutant mice lacking phosphorylation of myogenin at threonine 87. Mol. Cell. Biol. 24:1983-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J. C., and D. J. Goldhamer. 1999. Transcriptional mechanisms regulating MyoD expression in the mouse. Cell Tissue Res. 296:213-219. [DOI] [PubMed] [Google Scholar]
- 7.Choi, R. C., N. L. Siow, S. Q. Zhu, and K. W. Tsim. 2000. The cAMP-dependent protein kinase mediates the expression of AChE in chick myotubes. Neuroreport 11:801-806. [DOI] [PubMed] [Google Scholar]
- 8.Choi, R. C., N. L. Siow, S. Q. Zhu, D. C. Wan, Y. H. Wong, and K. W. Tsim. 2001. The cyclic AMP-mediated expression of acetylcholinesterase in myotubes shows contrasting activation and repression between avian and mammalian enzymes. Mol. Cell. Neurosci. 17:732-745. [DOI] [PubMed] [Google Scholar]
- 9.Choi, R. C., L. Y. Yung, T. T. Dong, D. C. Wan, Y. H. Wong, and K. W. Tsim. 1998. The calcitonin gene-related peptide-induced acetylcholinesterase synthesis in cultured chick myotubes is mediated by cyclic AMP. J. Neurochem. 71:152-160. [DOI] [PubMed] [Google Scholar]
- 10.DeChiara, T. M., D. C. Bowen, D. M. Valenzuela, M. V. Simmons, W. T. Poueymirou, S. Thomas, E. Kinetz, D. L. Compton, E. Rojas, J. S. Park, C. Smith, P. S. DiStefano, D. J. Glass, S. J. Burden, and G. D. Yancopoulos. 1996. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell 85:501-512. [DOI] [PubMed] [Google Scholar]
- 11.de Kerchove D'Exaerde, A., J. Cartaud, A. Ravel-Chapuis, T. Seroz, F. Pasteau, L. M. Angus, B. J. Jasmin, J. P. Changeux, and L. Schaeffer. 2002. Expression of mutant Ets protein at the neuromuscular synapse causes alterations in morphology and gene expression. EMBO Rep. 3:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, W. Q., M. Dong, D. Ninova, E. L. Holicky, M. D. Stegall, and L. J. Miller. 2003. Forskolin suppresses insulin gene transcription in islet beta-cells through a protein kinase A-independent pathway. Cell. Signal. 15:27-35. [DOI] [PubMed] [Google Scholar]
- 13.Duclert, A., and J. P. Changeux. 1995. Acetylcholine receptor gene expression at the developing neuromuscular junction. Physiol. Rev. 75:339-368. [DOI] [PubMed] [Google Scholar]
- 14.Fontaine, B., A. Klarsfeld, and J. P. Changeux. 1987. Calcitonin gene-related peptide and muscle activity regulate acetylcholine receptor alpha-subunit mRNA levels by distinct intracellular pathways. J. Cell Biol. 105:1337-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fromm, L., and S. J. Burden. 2001. Neuregulin-1-stimulated phosphorylation of GABP in skeletal muscle cells. Biochemistry 40:5306-5312. [DOI] [PubMed] [Google Scholar]
- 16.Glass, D. J., D. C. Bowen, T. N. Stitt, C. Radziejewski, J. Bruno, T. E. Ryan, D. R. Gies, S. Shah, K. Mattsson, S. J. Burden, P. S. DiStefano, D. M. Valenzuela, T. M. DeChiara, and G. D. Yancopoulos. 1996. Agrin acts via a MuSK receptor complex. Cell 85:513-523. [DOI] [PubMed] [Google Scholar]
- 17.Groisman, R., H. Masutani, M. P. Leibovitch, P. Robin, I. Soudant, D. Trouche, and A. Harel-Bellan. 1996. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J. Biol. Chem. 271:5258-5264. [DOI] [PubMed] [Google Scholar]
- 18.Gu, W., J. W. Schneider, G. Condorelli, S. Kaushal, V. Mahdavi, and B. Nadal-Ginard. 1993. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 72:309-324. [DOI] [PubMed] [Google Scholar]
- 19.Hai, T., and M. G. Hartman. 2001. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273:1-11. [DOI] [PubMed] [Google Scholar]
- 20.Heller, H., and E. Bengal. 1998. TFIID (TBP) stabilizes the binding of MyoD to its DNA site at the promoter and MyoD facilitates the association of TFIIB with the preinitiation complex. Nucleic Acids Res. 26:2112-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hribal, M. L., J. Nakae, T. Kitamura, J. R. Shutter, and D. Accili. 2003. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J. Cell Biol. 162:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, C. F., C. M. Neville, and J. Schmidt. 1993. Control of myogenic factor genes by the membrane depolarization/protein kinase C cascade in chick skeletal muscle. FEBS Lett. 319:21-25. [DOI] [PubMed] [Google Scholar]
- 23.Ip, F. C., D. G. Glass, D. R. Gies, J. Cheung, K. O. Lai, A. K. Fu, G. D. Yancopoulos, and N. Y. Ip. 2000. Cloning and characterization of muscle-specific kinase in chicken. Mol. Cell. Neurosci. 16:661-673. [DOI] [PubMed] [Google Scholar]
- 24.Kim, C. H., W. C. Xiong, and L. Mei. 2003. Regulation of MuSK expression by a novel signaling pathway. J. Biol. Chem. 278:38522-38527. [DOI] [PubMed] [Google Scholar]
- 25.Lacazette, E., S. Le Calvez, N. Gajendran, and H. R. Brenner. 2003. A novel pathway for MuSK to induce key genes in neuromuscular synapse formation. J. Cell Biol. 161:727-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, L., R. Heller-Harrison, M. Czech, and E. N. Olson. 1992. cAMP-dependent protein kinase inhibits the activity of myogenic helix-loop-helix proteins. Mol. Cell. Biol. 12:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, W., R. W. Burgess, B. Dominguez, S. L. Pfaff, J. R. Sanes, and K. F. Lee. 2001. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature 410:1057-1064. [DOI] [PubMed] [Google Scholar]
- 28.Luo, Z., Q. Wang, J. Zhou, J. Wang, M. Liu, X. He, A. Wynshaw-Boris, W. Xiong, B. Lu, and L. Mei. 2002. Regulation of AChR clustering by Dishevelled interacting with MuSK and PAK1. Neuron 35:489-505. [DOI] [PubMed] [Google Scholar]
- 29.Ma, L., Y. Z. Huang, G. M. Pitcher, J. G. Valtschanoff, Y. H. Ma, L. Y. Feng, B. Lu, W. C. Xiong, M. W. Salter, R. J. Weinberg, and L. Mei. 2003. Ligand-dependent recruitment of the ErbB4 signaling complex into neuronal lipid rafts. J. Neurosci. 23:3164-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, P. C., M. A. Rould, H. Weintraub, and C. O. Pabo. 1994. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77:451-459. [DOI] [PubMed] [Google Scholar]
- 31.Magenta, A., C. Cenciarelli, F. De Santa, P. Fuschi, F. Martelli, M. Caruso, and A. Felsani. 2003. MyoD stimulates RB promoter activity via the CREB/p300 nuclear transduction pathway. Mol. Cell. Biol. 23:2893-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11:497-504. [DOI] [PubMed] [Google Scholar]
- 33.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 34.Montminy, M. R., K. A. Sevarino, J. A. Wagner, G. Mandel, and R. H. Goodman. 1986. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc. Natl. Acad. Sci. USA 83:6682-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muscat, G. E., S. Rea, and M. Downes. 1995. Identification of a regulatory function for an orphan receptor in muscle: COUP-TF II affects the expression of the myoD gene family during myogenesis. Nucleic Acids Res. 23:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedraza-Alva, G., J. M. Zingg, and J. P. Jost. 1994. AP-1 binds to a putative cAMP response element of the MyoD1 promoter and negatively modulates MyoD1 expression in dividing myoblasts. J. Biol. Chem. 269:6978-6985. [PubMed] [Google Scholar]
- 37.Puri, P. L., M. L. Avantaggiati, C. Balsano, N. Sang, A. Graessmann, A. Giordano, and M. Levrero. 1997. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16:369-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puri, P. L., and V. Sartorelli. 2000. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J. Cell. Physiol. 185:155-173. [DOI] [PubMed] [Google Scholar]
- 39.Sanes, J. R., and J. W. Lichtman. 1999. Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 22:389-442. [DOI] [PubMed] [Google Scholar]
- 40.Sanes, J. R., and J. W. Lichtman. 2001. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat. Rev. Neurosci. 2:791-805. [DOI] [PubMed] [Google Scholar]
- 41.Sapru, M. K., S. K. Florance, C. Kirk, and D. Goldman. 1998. Identification of a neuregulin and protein-tyrosine phosphatase response element in the nicotinic acetylcholine receptor e subunit gene: regulatory role of an ets transcription factor. Proc. Natl. Acad. Sci. USA 95:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaeffer, L., A. de Kerchove d'Exaerde, and J. P. Changeux. 2001. Targeting transcription to the neuromuscular synapse. Neuron 31:15-22. [DOI] [PubMed] [Google Scholar]
- 43.Schaeffer, L., N. Duclert, M. Huchet-Dymanus, and J.-P. Changeux. 1998. Implication of a multisubunit Ets-related transcription factor in synaptic expression of the nicotinic acetylcholine receptor. EMBO J. 17:3078-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 45.Si, J., Z. Luo, and L. Mei. 1996. Induction of acetylcholine receptor gene expression by ARIA requires activation of mitogen-activated protein kinase. J. Biol. Chem. 271:19752-19759. [DOI] [PubMed] [Google Scholar]
- 46.Si, J., and L. Mei. 1999. ERK MAP kinase activation is required for ARIA-induced increase in all five AChR subunit mRNAs as well as synapse-specific expression of the AChR e-transgene. Mol. Brain Res. 67:18-27. [DOI] [PubMed] [Google Scholar]
- 47.Spinner, D. S., S. Liu, S. W. Wang, and J. Schmidt. 2002. Interaction of the myogenic determination factor myogenin with E12 and a DNA target: mechanism and kinetics. J. Mol. Biol. 317:431-445. [DOI] [PubMed] [Google Scholar]
- 48.Tang, H., P. Macpherson, L. S. Argetsinger, D. Cieslak, S. T. Suhr, C. Carter-Su, and D. Goldman. 2004. CaM kinase II-dependent phosphorylation of myogenin contributes to activity-dependent suppression of nAChR gene expression in developing rat myotubes. Cell. Signal. 16:551-563. [DOI] [PubMed] [Google Scholar]
- 49.Tansey, M. G., G. C. Chu, and J. P. Merlie. 1996. ARIA/HRG regulates AChR epsilon subunit gene expression at the neuromuscular synapse via activation of phosphatidylinositol 3-kinase and Ras/MAPK pathway. J. Cell Biol. 134:465-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valenzuela, D. M., T. N. Stitt, P. S. DiStefano, E. Rojas, K. Mattsson, D. L. Compton, L. Nunez, J. S. Park, J. L. Stark, D. R. Gies, S. Thomas, M. M. Le Beau, A. A. Fernald, N. G. Copeland, N. A. Jenkins, S. J. Burden, D. J. Glass, and G. D. Yancopoulos. 1995. Receptor tyrosine kinase specific for the skeletal muscle lineage: expression in embryonic muscle, at the neuromuscular junction, and after injury. Neuron 15:573-584. [DOI] [PubMed] [Google Scholar]
- 51.Walton, K. M., R. P. Rehfuss, J. C. Chrivia, J. E. Lochner, and R. H. Goodman. 1992. A dominant repressor of cyclic adenosine 3′,5′-monophosphate (cAMP)-regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol. Endocrinol. 6:647-655. [DOI] [PubMed] [Google Scholar]
- 52.West, A. E., W. G. Chen, M. B. Dalva, R. E. Dolmetsch, J. M. Kornhauser, A. J. Shaywitz, M. A. Takasu, X. Tao, and M. E. Greenberg. 2001. Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. USA 98:11024-11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, X., S. Arber, C. William, L. Li, Y. Tanabe, T. M. Jessell, C. Birchmeier, and S. J. Burden. 2001. Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron 30:399-410. [DOI] [PubMed] [Google Scholar]
- 54.Yuan, W., G. Condorelli, M. Caruso, A. Felsani, and A. Giordano. 1996. Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem. 271:9009-9013. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, Q., R. W. Gedrich, and D. A. Engel. 1995. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J. Virol. 69:4323-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]