FIGURE 1.
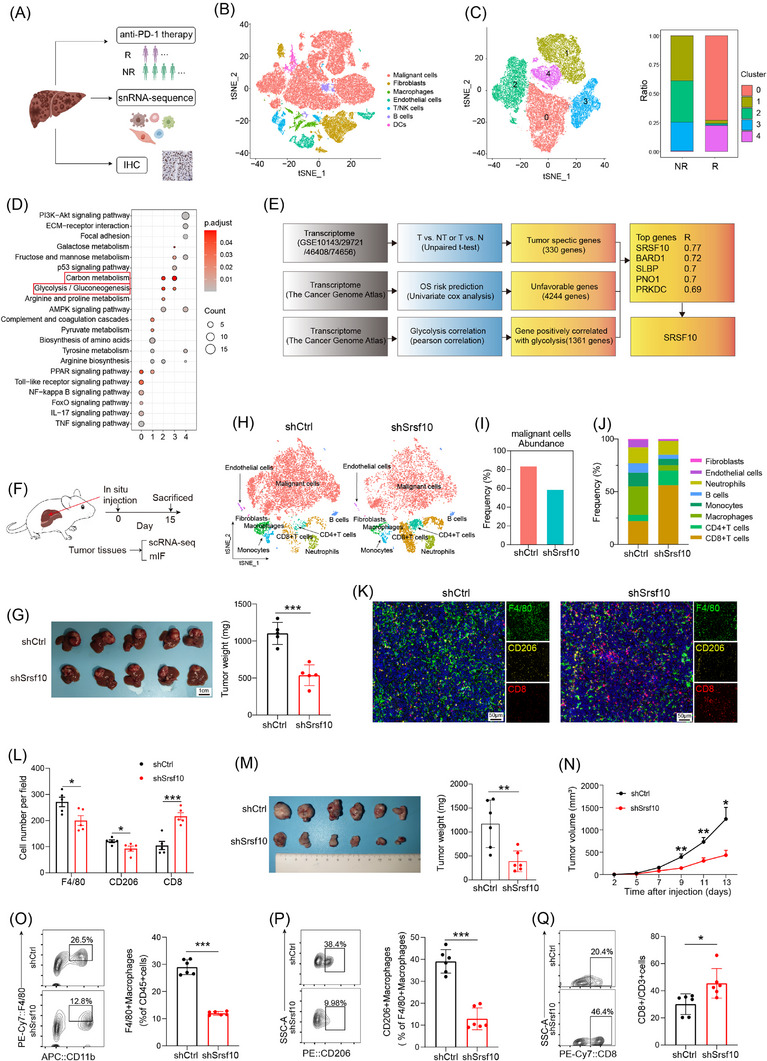
SRSF10 was a crucial gene associated with anti‐PD‐1 resistance and immunosuppressive microenvironment with HCC. (A) Schematic diagram of the recruitment of patients and specimens in the training cohort. (B‐C) t‐SNE analysis of snRNA‐seq data from HCC tumor tissues. (D) KEGG analysis of genes associated with tumor cells from snRNA‐seq. (E) Model diagram for screening of glycolysis‐related genes. (F) Workflow of the HCC model in situ. (G) Tumor picture and statistical analysis of orthotopic HCC model (n = 6). (H) t‐SNE analysis of scRNA‐seq data from mouse HCC tumor tissues. (I) Comparative abundance of the cancer cells in control versus Srsf10‐knockdown HCC tumors, extracted from the scRNA‐seq data. (J) Comparative abundance of the tumor microenvironment in control versus Srsf10‐knockdown HCC tumors, extracted from the scRNA‐seq data. (K) Representative pictures of mIF analysis for F4/80, CD206 and CD8 markers (n = 5). (L) Quantification of corresponding immune cells by mIF analysis (n = 5). (M) Representative image and statistical tests of a Hepa1‐6 cell‐derived tumor harvested from a C57BL/6 mouse on day 14 (n = 6). (N) Tumor growth curves of subcutaneous HCC tumor (n = 6). (O‐Q) Representative plots and percentages of tumor‐infiltrating F4/80+ macrophages, CD206+ macrophages and CD8+ T cells from control and shSrsf10 HCC tumor (n = 6). Two‐tailed unpaired Student's t test (G, L ‐Q); ns, not significant; * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: IHC, Immunohistochemistry; DC, Dendritic cell; NR, non‐response; SRSF10, serine and arginine rich splicing factor 10; BARD1, BRCA1 associated RING domain 1; SLBP, stem‐loop histone mRNA binding protein; PNO1, partner of NOB1 homolog; PRKDC, protein kinase, DNA‐activated, catalytic subunit; OS, overall survival; scRNA‐seq, single‐cellar RNA sequencing; mIF, multicolor immunofluorescence; tSNE, t‐distributed Stochastic Neighbor Embedding; HCC, hepatocellular carcinoma; snRNA‐seq, snRNA‐seq: single‐nuclear RNA sequencing.
