FIGURE 5.
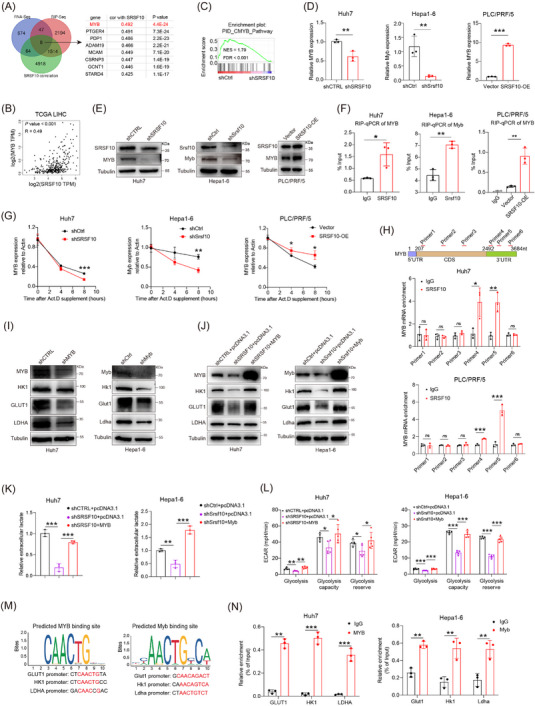
SRSF10 positively regulated glycolysis in HCC cells through transcription factor MYB. (A) Venn diagram was used to identify the overlapping genes in indicated data. (B) Correlation of SRSF10 and MYB in TCGA‐LIHC dataset. (C) GSEA analysis of CMYB pathway enriched in differential gene from RNA‐seq. (D) MYB mRNA levels in Huh7, PLC/PRF/5, and Hepa1‐6 cells, detected by qRT‐PCR analyses (n = 3). (E) Western blotting was performed in Huh7, PLC/PRF/5, and Hepa1‐6 cells. (F) RIP assays to verify the binding of SRSF10 and MYB in Huh7, PLC/PRF/5, and Hepa1‐6 cells (n = 3). (G) Huh7, PLC/PRF/5, and Hepa1‐6 cells were treated with actinomycin D as indicated times. The mRNA expression levels of MYB were examined using qRT‐PCR. Error bars are mean ± SD from three biologically independent samples (n = 3). (H) CLIP assays to verify the binding region of SRSF10 and MYB in Huh7 and PLC/PRF/5 cells (n = 3). (I) Western blotting was performed in Huh7 and Hepa1‐6 cells. (J) Western blotting was performed in control, shSRSF10, and shSRSF10 followed by MYB overexpression in Huh7 and Hepa1‐6 cells (n = 3). (K) Detection of lactate content in control, shSRSF10, and shSRSF10 followed by MYB overexpression in Huh7 and Hepa1‐6 cells. (L) ECAR of huh7 cells and hepa1‐6 cells with control, shSRSF10, and shSRSF10 followed by MYB overexpression groups (n = 5). (M) Predicted binding sites of the transcription factor MYB within the promoter sequences of GLUT1, HK1, and LDHA. (N) MYB binding to the GLUT1, HK1, LDHA promoter was determined by chromatin immunoprecipitation‐RT‐PCR in Huh7 and Hepa1‐6 cells (n = 3). Two‐tailed unpaired Student's t test (D, F‐H, K‐N); ns, not significant; * P < 0.05, ** P < 0.01, *** P < 0.001. Abbreviations: MYB, MYB proto‐oncogene, transcription factor; PTGER4, prostaglandin E receptor 4; PDP1, pyruvate dehydrogenase phosphatase catalytic subunit 1; ADAM19, ADAM metallopeptidase domain 19; MCAM, melanoma cell adhesion molecule; CSRNP3, cysteine and serine rich nuclear protein 3; GCNT1, glucosaminyl (N‐acetyl) transferase 1; STARD4, StAR related lipid transfer domain containing 4; RNA‐seq, RNA Sequencing; RIP‐seq, RNA Immunoprecipitation Sequencing; NES, Normalized enrichment score; FDR, False Discovery Rate; SRSF10, serine and arginine rich splicing factor 10; OE, over expression; TCGA, The Cancer Genome Atlas; LIHC, Liver hepatocellular carcinoma; Act D, actinomycin D; 5’UTR, 5' Untranslated Regions; 3’UTR, 3' Untranslated Regions; CDS, Coding sequence; HK1, hexokinase 1; GLUT1, Glucose transporter 1; LDHA, lactate dehydrogenase A; GSEA, Gene Set Enrichment Analysis; qRT‐PCR, Quantitative Real‐Time Reverse Transcription Polymerase Chain Reaction; CLIP, cross‐linking and immune‐precipitation; ECAR, extracellular acidification rate.
