Abstract
The human mitochondrial tRNALeu(CUN) [hmtRNALeu(CUN)] corresponds to the most abundant codon for leucine in human mitochondrial protein genes. Here, in vitro studies reveal that the U48C substitution in hmtRNALeu(CUN), which corresponds to the pathological T12311C gene mutation, improved the aminoacylation efficiency of hmtRNALeu(CUN). Enzymatic probing suggested a more flexible secondary structure in the wild-type hmtRNALeu(CUN) transcript compared with the U48C mutant. Structural analysis revealed that the flexibility of hmtRNALeu(CUN) facilitates a T-stem slip resulting in two potential tertiary structures. Several rationally designed tRNALeu(CUN) mutants were generated to examine the structural and functional consequences of the T-stem slip. Examination of these hmtRNALeu(CUN) mutants indicated that the T-stem slip governs tRNA accepting activity. These results suggest a novel, self-regulation mechanism of tRNA structure and function.
INTRODUCTION
In protein biosynthesis, transfer RNAs (tRNAs) play a central role in gene expression as adaptor molecules of the codons in mRNA and amino acids (1). The human mitochondrial translation machinery is dependent on 22 tRNAs, one for each of 18 amino acids and two for Leu and Ser with different anticodons (2). All of these tRNAs are encoded by the mitochondrial genome. The primary and secondary structures of human mitochondrial tRNAs (hmtRNAs) differ significantly from those of canonical bacterial and cytoplasmic tRNAs, and tRNAs in human mitochondria are less thermodynamically stable because they generally contain higher numbers of mismatched and AU base pairs (3). Therefore, while hmtRNAs should adopt an L-shape tertiary structure of canonical tRNA in order to function in ribosomal protein synthesis, their folded structures may be constructed with different sets of intramolecular contacts that are mostly unknown.
In the past 15 years, a number of point mutations in hmtRNA genes have been found to be correlated with a variety of multi-system diseases (4,5). Although the molecular mechanisms of these mitochondrial DNA-mediated diseases remain unclear, accumulating evidence has shown that severe structural and functional defects of hmtRNAs are caused by the pathogenic mutations (6–8). Systematic investigation of the structure and function of hmtRNAs can, therefore, provide valuable information about related diseases and potentially facilitate development of diagnostic tools and therapies for these diseases.
Among the 22 hmtRNAs, hmtRNALeu(CUN) corresponds to the most frequently used codon (14.9%) (9). Even a slight impairment of the function of hmtRNALeu(CUN) can lead to significant deficiencies in mitochondrial protein synthesis. Although tRNALeu(CUN) is one of the few mitochondrial tRNAs that possesses all of the structural features for a classical cloverleaf structure and 3D folding (3), little information is available about the structure and function of hmtRNALeu(CUN).
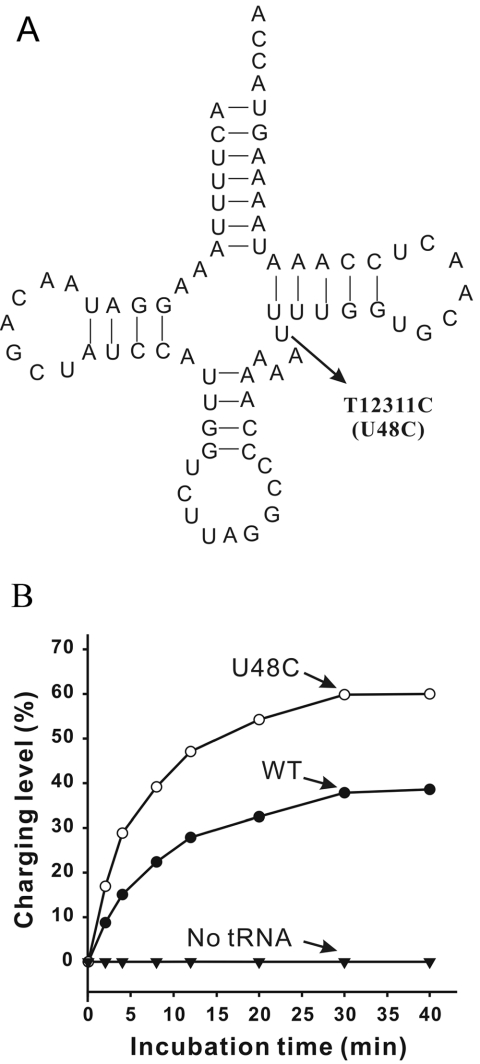
Among the five known pathogenic mutations in the hmtRNALeu(CUN) gene (see http://www.mitomap.org) is the T12311C mutation (10) that sparks our interest. This mutation resides at residue 48, which is the connector between the variable loop and the T-stem in the tRNA secondary structure (Figure 1A). It is hypothesized that the tertiary interaction between nt 15 and 48 plays an important role in the tRNA 3D structure; replacement of either of these 2 nt affects tRNA conformation (11). In this in vitro study, a U48C substitution was introduced in the hmtRNALeu(CUN) gene, mimicking the T12311C mutation, in order to examine changes in structure and aminoacylation of hmtRNALeu(CUN). Surprisingly, the tRNA accepting capacity and structural stability were increased by this substitution. Secondary structure analysis of hmtRNALeu(CUN) suggested two kinds of pairing alignments in the T-stem that could be formed by a 1 nt slip. We subsequently constructed a series of hmtRNALeu(CUN) mutants to mimic the different types of tertiary structures resulting from the T-stem slip and studied their structures and aminoacylation capacities. Here, we analyze the structural basis of T-stem slip and the resulting tertiary structures that provide evidence for a novel, self-regulating acceptance mechanism of hmtRNALeu(CUN).
Figure 1.
The effect of pathogenic T12311C (U48C) mutation on the aminoacylation hmtRNALeu(CUN). (A) Theoretical cloverleaf structure of tRNALeu(CUN) as deduced from the DNA sequence. The pathogenic point mutation T12311C (U48C) is indicated with arrow. (B) Aminoacylation of WT hmtRNALeu(CUN) and U48C mutant transcripts.
MATERIALS AND METHODS
Enzyme purification and tRNA preparation
All chemicals were purchased from Sigma–Aldrich Inc. (USA), except otherwise noted.
T7 RNA polymerase and human mitochondrial leucyl-tRNA synthetase (hmLeuRS) were purified from Escherichia coli overproducing strains as described previously in our laboratory (12,13). In vitro transcription of mitochondrial tRNA and subsequent refolding of tRNA were performed as described previously (14). The tRNA concentration was determined by UV absorbance at 260 nm and the extinction coefficient was calculated from the sequence of each tRNA (15).
Assaying aminoacylation of tRNA transcript using [14C]leucine
Accepting activity of the tRNA transcript and the time course of tRNA aminoacylation were assessed as described previously (14). The optimized reaction mixture contained 50 mM HEPES, pH 7.6, 25 mM KCl, 10 mM MgCl2, 2.5 mM ATP, 1 mM spermidine, 100 μg/ml BSA, 20 μM L-[14C]leucine, 0.5 μM hmLeuRS and 1.5 μM hmtRNALeu(CUN) transcript. The aminoacylation plateau was determined during a 40 min incubation time. The charging level of each tRNA transcript was calculated as the percentage of aminoacylated tRNA versus the total amount of specific tRNA per experiment. The apparent kinetic parameters, kcat and KM, of hmLeuRS for hmtRNALeu(CUN) and its mutants were derived from Lineweaver-Burk plots on experiments performed with 1–50 μM tRNA and 0.15 μM hmLeuRS. Displayed data were the averages of three independent experiments.
Secondary structure prediction of tRNA
MFOLD 3.1 program (http://www.bioinfo.rpi.edu/~zukerm/) was used to analyze the secondary structure of tRNA (16). Parameters were used in default settings, except for percent suboptimality, which was set as 50%. Only cloverleaf-like structures were considered.
Nuclease mapping of tRNA structure in solution
tRNA transcripts were 5′-end-labeled with 32P by the reaction with T4 polynucleotide kinase (New England Biolabs, Canada) and [γ-32P]ATP as described previously (17). The purified 32P-labeled transcripts were denatured by heating to 60°C in the absence of Mg2+ and then cooled slowly in the presence of 10 mM MgCl2 (18). 32P-labeled tRNAs were digested with various nucleases at 25°C for 10 min in 20 μl of 40 mM Tris–HCl, pH 7.5, 10 mM MgCl2 and 40 mM NaCl. For digestion with nuclease S1 (Amersham Pharmacia, UK), 1 mM ZnCl2 was added. The digestion reaction mixtures contained 10 000 c.p.m. 32P-labeled tRNA, 4 μg non-labeled carrier tRNA and 0.1 or 2.0 U RNase T1 (Amersham, USA), 0.035 or 0.1 U RNase V1 (Ambion, USA) and 50 or 150 U nuclease S1, respectively. Reactions were stopped by adding 8 μg of carrier tRNA, chilling on ice and phenol extraction. The digested products were recovered by ethanol precipitation. The pellets were resuspened in 10 μl of dye mix solution [0.0125% xylene cyanol and 0.0125% bromphenol blue in 50% formamide]. Samples were run on a 21 cm (width) × 50 cm (height) × 0.4 mm (diameter) 12% polyacrylamide gel containing 8 M urea buffered with 1× TBE, pH 8.3). Alkaline ladders and G-ladders were obtained as described previously (19).
RESULTS
Aminoacylation capacity of tRNALeu(CUN) was increased by the pathogenic T12311C (U48C) mutation
In the hmtRNALeu(CUN) gene, the pathogenic T12311C mutation changes the U to C at position 48, which is the connecting base between the variable loop and the T-stem (Figure 1A). We constructed the wild-type (WT) hmtRNALeu(CUN) and the U48C mutant in vitro and tested their aminoacylation efficiency by the cognate hmLeuRS.
Interestingly, the accepting activity of the mutant tRNA with the U48C substitution was increased compared with the WT (Figure 1B). Under optimal conditions, 40% of the WT transcript was aminoacylated (compared with the calculated theoretical accepting activity of the transcript, see Materials and Methods), while the accepting activity of the U48C mutant was >60%. Kinetic aminoacylation assays were further performed with 1–50 μM tRNA and 0.15 μM hmLeuRS. The kinetic parameters kcat and KM were determined for both the mutant and WT transcripts. Establishment of these parameters enables a valuable comparison of aminoacylation efficiency (kcat/KM). The increase of charging capacity was further evidenced by the finding that the U48C mutant was leucylated with a 1.31-fold higher efficiency than the WT hmtRNALeu(CUN) (Table 1).
Table 1.
Apparent kinetic parameters of in vitro aminoacylation of WT and mutant hmtRNALeu(CUN) transcripts by human mitochondrial leucyl-tRNA synthetase
| hmtRNALeu(CUN) transcripts | kcat(10−3 s−1) | KM(μM) | kcat/KM(10−3 s−1 μM−1) | Relative kcat/KM |
|---|---|---|---|---|
| Wild type | 52.4 ± 1.8 | 23.9 ± 3.9 | 2.19 | 1 |
| U48C | 112.3 ± 1.0 | 39.1 ± 5.7 | 2.87 | 1.31 |
| U48G | 115.5 ± 3.2 | 36.3 ± 6.9 | 3.18 | 1.45 |
| U48A | 110.3 ± 2.2 | 37.2 ± 5.3 | 2.96 | 1.35 |
| Δ48 | 123.1 ± 4.7 | 34.9 ± 2.6 | 3.53 | 1.61 |
| G52A/C62G | 53.8 ± 3.2 | 11.5 ± 2.5 | 4.68 | 2.13 |
| U51A/A63U | 56.2 ± 4.9 | 8.8 ± 1.6 | 6.39 | 2.91 |
| U51G | <0.0001 | Nd | – | – |
| U48C/+G54 | <0.0001 | Nd | – | – |
Aminoacylation reactions were as described in Materials and Methods. Data presented here were the average of three independent experiments.
Nd, not determined.
Because magnesium ions may affect tRNA folding, we assayed the denaturing/renaturing processes with several concentrations of magnesium (from 3 to 20 mM) for each transcript. Similar aminoacylation levels of WT and mutant tRNA were obtained under all of these conditions (data not shown), indicating that magnesium ion concentration had little effect on hmtRNALeu(CUN) folding.
Structure analysis of hmtRNALeu(CUN)
In order to understand the relatively lower aminoacylation efficiency of the WT transcript, we examined the secondary structure of hmtRNALeu(CUN) using the MFOLD program (16) and enzymatic probing (20).
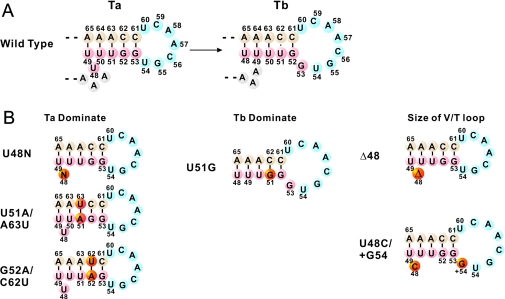
Secondary structure calculation revealed that there were two types of T-stem base pair alignments in the hmtRNALeu(CUN) transcript. In one alignment (Ta alignment), the U48 was located between the T-stem and variable loop as an unpaired connector and the T-loop contained 7 nt. In the other alignment (Tb alignment), the U48 paired with A65 in the T-stem and the enlarged T-loop contained 8 nt. Thus, the Ta alignment could become the Tb alignment by a 1 nt slip, which formed a base pair between 48 and 65 bases (Figure 2A). Because the U48C substitution decreased the tendency to form a base pair between C48 and A65, thus prohibiting the T-stem slip, most of the U48C transcript was stabilized in Ta alignment.
Figure 2.
Predicated T-stem secondary structures of WT hmtRNALeu(CUN) and the designed variants. Numbering of the nucleotides is according to Sprinzl et al. (39). (A) Two alternative T-stem alignments in WT hmtRNALeu(CUN) transcript. U48 at the variable loop of Ta alignment slips to T-stem and forms a new U48:A65 base pair in Tb alignment with 8 nt T-loop; (B) various hmtRNALeu(CUN) constructs with designed T-stem. The U48N (N = A, G, C), G52A/C62U and U51A/A63U mutants were dominated in the Ta alignment; the U51G mutation was dominated in the Tb alignment; the Δ48 and U48C/+G54 mutants were designed to mimic the effect of T-stem slip on V-loop and T-loop, respectively.
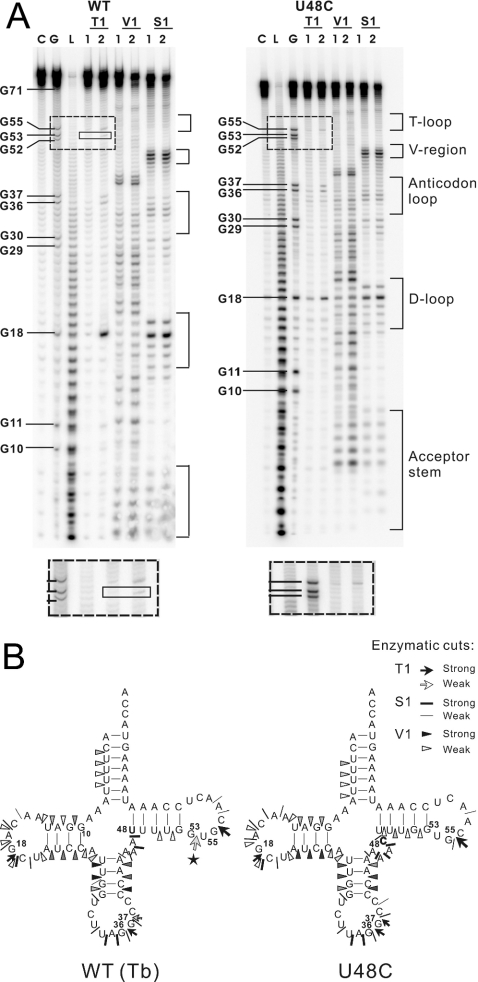
Considering the limitations of the MFOLD prediction (e.g. its ignorance of RNA 3D structure), we carried out an enzymatic probing experiment to detect the solution structure of the WT and mutant transcripts. The results of the enzymatic probing of hmtRNALeu(CUN) were in agreement with the MFOLD prediction described above. Nuclease S1 and RNase T1 (guanine specific) were used to probe the unpaired nucleotides, and RNase V1 was used to monitor base-paired or stacked nucleotides. The 5′-end-labeled cleaved fragments were separated by PAGE/urea. As the nonspecific cleavage of the transcripts at certain weak points could affect the results, each of these enzymes was used with different concentrations. Only dose-dependent enzymatic cleavages were considered. The autoradiograms are presented in Figure 3A and results of enzymatic mapping are summarized in Figure 3B.
Figure 3.
Comparative enzymatic probing of in vitro transcribed human mitochondrial tRNALeu(CUN) and the U48C mutant. (A) Autoradiograms of the various cleavage products of 5′-end-labeled tRNA transcripts separated on denaturing 12% polyacrylamide gels. Lane C, control incubations without probe; lane L, alkaline ladder; lane G, G ladder. The tRNA was 5′-labeled and digested with RNase T1, RNase V1 and nuclease S1. Numbers 1 and 2 refer to increasing concentrations of nuclease. The RNase T1 cleavage product specific for WT transcript was indicated in the solid-line square. The difference can be easily seen in the dashed-line squares. (B) Result of the enzymatic probing of WT and U48C mutant hmtRNALeu(CUN) transcripts. Intensities of cuts are proportional to the darkness of the symbols. The pentacle denotes the specific RNase T1 cleavage at G53 on WT transcript.
Most of the nucleotides within the theoretical stem regions of the WT hmtRNALeu(CUN) were cleaved by RNase V1 and most of those within the canonical loop regions were recognized by nuclease S1. The hmtRNALeu(CUN) transcript appears to fold into the classical cloverleaf structure despite the absence of post-transcriptional modification. The general cleavage patterns of the WT and U48C mutant were similar to each other except for some minor differences in their T-loops. The WT transcript was cleaved by RNase T1 at G53, which is thought to be paired with C61 in the Ta alignment. This indicated that at least some proportion of the WT transcript existed in the Tb alignment, in which G53 remained unpaired in the T-loop. The cleavage at G53 by RNase T1 was dose-dependent in three separate experiments (data not shown), suggesting that the result is not related to the quality of the gel. However, in the case of U48C mutant, no corresponding cleavage was observed. The differences in the enzymatic digestion patterns of the WT and U48C mutant are summarized in Figure 3B. It appears that the fragment indicated with a pentacle in Figure 3B was due to the U48 shift in the T-stem.
Manipulation of the T-stem slip of hmtRNALeu(CUN)
To investigate the effect of the different T-stem base pair alignments on the structure and aminoacylation of hmtRNALeu(CUN), we constructed mutants of hmtRNALeu(CUN) that would stabilize in either the Ta or Tb alignment depending on the T-stem structure (Figure 2B). Similar to U48C mutant, U48A and U48G mutants were created to restrain the T-stem slip by prohibiting the base pairing between 48 and 65 residues. These two mutants would theoretically stabilize in the Ta alignment. Two double mutations, U51A/A63U and G52A/C62U, were designed to prevent the T-stem from slipping by introducing an inside bolt in the T-stem. In the U51A/A63U mutant, the U50•A63 base pair became unpaired U50•U63 so that the Tb alignment of the T-stem was unfavorable and the Ta alignment dominated. Similarly, in G52A/C62U mutant the unpaired U51•U62 was unstable, favoring the Ta alignment of T-stem. Another mutant, U51G, was constructed to facilitate a T-stem slip by forming a base pair between G51 and C62, thus favoring a Tb alignment. In the U51G mutant, the T-stem slip reduced the size of the variable loop 1 nt (from 4 to 3) and enlarged the T-loop by 1 nt (from 7 to 8) relatively. To ascertain the effect of the alteration on aminoacylation of hmtRNALeu(CUN), mutants were constructed with a 3 nt variable loop or 8 nt T-loop. The Δ48 mutant had a deleted nucleotide at position 48 to test the effect of a reduced variable loop and the U48C/+G54 mutant contained 8 nt in its T-loop.
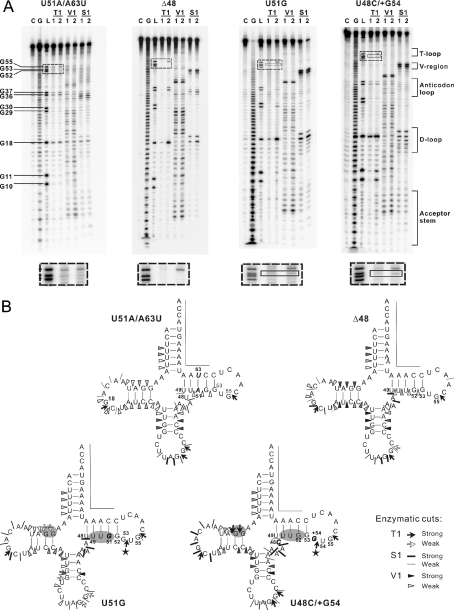
The design for each tRNA variant was confirmed by secondary structure prediction (data not shown). Enzymatic probing was further performed with the representative tRNA transcripts. Although the digestion signal at the T-loop was weak, probably due to the tertiary structure protection, the expected alignment patterns were still detectable. As presented in Figure 4A, no dose-dependent RNase T1 cleavage site at G53 was detected in either the U51A/A63U or the Δ48 mutant, which indicated that the T-stem slip was restrained by these mutations. Meanwhile in the U51G and U48C/+G54 mutants, RNase T1 cleaved at G53 and G54, respectively, which was consistent with the T-loop enlargement. Moreover, some other characteristic differences in the structures of tRNA variants were detected. For example, the nucleotides in the T-stem were cleaved by RNase V1 in the 7 nt T-loop variant (Δ48, U48C and U51A/A63U), whereas T-stem cleavages did not occur in either of the mutants with an enlarged T-loop (U51G and U48C/+G54). Moreover, G10 and G11 in the D-stems of both the U51G and U48C/+G54 mutants were cleaved by RNase T1, suggesting that the T-loop enlargements induced changes in their D-loops. These differences most likely reflect domain–domain interactions between the D- and T-loops in hmtRNALeu(CUN). The patterns of enzymatic digestion of each tRNA mutant are given in Figure 4B.
Figure 4.
Enzymatic probing of the representative tRNALeu(CUN) variants. (A) Cleavage products of 5′-labeled molecules after treatment with nucleases, displayed on autoradiograms of 12% polyacrylamide gels. C stands for H2O ladder; G for G ladder; L for alkaline ladder; T1 for RNase T1 digestion ladder; V1 for RNase V1 digestion ladder; S1 for nuclease S1 digestion ladder; numbers 1 and 2 refer to increasing concentrations of nuclease. The RNase T1 cleavage product denoted in the solid-line square indicates the enlarged T-loop. The band difference can be easily seen in the dashed-line squares. (B) Location of enzymatic cleavage sites on cloverleaf diagrams of the transcripts. Specifications and intensities of cuts are as indicated in the key. Nucleotides that could not be tested because of technical limitations are marked by a line. Mutated nucleotides are shown in italics. Regions with different cleavage pattern are highlighted by gray background. The pentacles denote the RNase T1 cleavage specific for the enlarged T-loop.
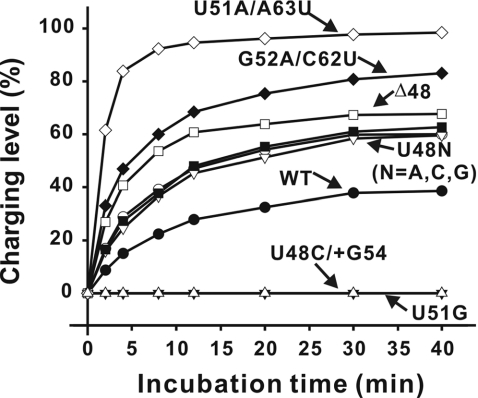
The charging efficiency of hmLeuRS for each designed tRNA variant was examined. The accepting activities of these mutants are shown in Figure 5 and the kinetic parameters are presented in Table 1. The aminoacylation of the three U48N (N = A, G, C) mutants increased similarly compared with the WT transcript. The results indicate that the increased accepting activity of the U48C mutation is due to the secondary structure rather than a specific residue substitution. The two T-stem double mutants showed greater increases (2.91-fold for U51A/A63U and 2.13-fold for G52A/C62U as compared with that for WT transcript) in charging efficiency (kcat/KM) than those of U48 mutants, suggesting that the stability of Ta alignment and instability of Tb alignment is very important for the accepting activity. The Δ48 mutant, which mimicked the shortened variable loop resulting from a T-stem slip, also presented increased plateau level (65%) and charging efficiency (1.61-fold increase relative to WT). It is possible that the shortened variable loop inhibited T-stem slip, favoring the Ta alignment. Conversely, the U51G mutation that promoted the T-stem slip, favoring the Tb alignment, dramatically decreased the charging activity of hmtRNALeu(CUN). Less than 0.5% of the U51G mutant was aminoacylated following a 40 min incubation under the experimental conditions. The U48C/+G54 mutation with the 8 nt T-loop, which mimicked the T-loop induced by a T-stem slip, also showed decreased aminoacylation. Neither of these two mutants displayed sufficient aminoacylation to determine the kinetic parameters. This result suggests that the size of the T-loop is critical for the tRNA charging activity in that charging capacity is decreased by the addition of bases into the T-loop, even when T-stem is unchanged. The data suggest that the negative effect of a T-stem slip on aminoacylation of hmtRNALeu(CUN) is due to the increased size of the T-loop induced by the slip.
Figure 5.
Effects of the designed mutations on aminoacylation capacity of hmtRNALeu(CUN). All values were the average of three experiments. The standard errors were <5%.
DISCUSSION
Comparison of the variable loops and T-stems of E.coli (21), Aquifex aeolicus (22) and human cytoplasmic (23) tRNALeus with those of hmtRNALeu(CUN) indicates that there are several features that determine whether a tRNALeus structure is conducive to a T-stem slip. All 5 bp in the T-stem of E.coli tRNALeu(CUG) and A.aeolicus tRNALeu(CUC) and the 4 bp in the T-stem of E.coli tRNALeu(CUC) and human cytoplasmic tRNALeu(CUU) are GC pairs. This high level of GC base pairing in the T-stem stabilizes the Ta alignment and thereby inhibits a T-stem slip in the above tRNALeus. A T-stem slip is further disabled by the inability of the base at position 48 in the variable loop to form a base pair with the base at position 65 in these tRNALeus. Finally, the large size of the variable loops in these Ta stable tRNALeus (12–16 nt and 2–4 bp) is a hindrance to a T-stem slip. In contrast, because only 2 of the 5 bp of the T-stem are GC pairs in hmtRNALeu(CUN), the Ta structure configuration is less stable than that in the above tRNAs. Moreover, in hmtRNALeu(CUN) U48 exactly pairs with A65 following a T-stem slip, and the variable loop is smaller containing only 4 nt and no base pair. These unique structural features of hmtRNALeu (CUN) enable a T-stem slip.
The inherently fragile structure of hmtRNA has been discussed previously (24–26). In this work, the structural fragility of hmtRNALeu(CUN) is specified as a mechanism of T-stem slip. The T-stem slip can result in a mixture of Ta and Tb alignments among the WT hmtRNALeu(CUN). Mutation analysis indicates that the T-stem slip, especially the varied size of the T-loop, correlates with the charging capacity. When its T-stem presents a Ta alignment and the T-loop contains 7 nt, the tRNA can be charged. While the T-stem slips to the Tb alignment and the T-loop is enlarged to 8 nt, the accepting capacity is lost. The T-stem slip appears to function as a switch for the charging capacity of hmtRNALeu(CUN).
As the T-stem is generally not a locus for tRNALeus recognition in systems including E.coli, Haloferax volcanii, Saccharomyces cerevisiae and human mitochondria (27–32), it is highly probable that the T-stem slip in hmtRNALeu(CUN) governs its charging capacity by inducing a change in tRNA tertiary structure. Enzymatic probing experiments reveal the D-stem relaxation in mutants with an enlarged T-loop (Figure 4), indicating domain–domain interactions between the D- and T-loops in hmtRNALeu(CUN). Considering the interdomain interactions found in other hmtRNAs (25,26,33), the prevalence of intramolecular communication could be expected in the tRNA possessing a fragile structure.
The generation of a functional tRNA is a complicated process with many steps, including transcription, end-processing, post-transcriptional modification, aminoacylation and participation in ribosomal protein synthesis. Previous work has shown that a tRNA molecule can assume different conformations to interact with different partners (34–36). The alternative conformations of hmtRNALeu(CUN) that result from the T-stem slip may be required for its optimal interaction with the processing enzymes or the translation apparatus in human mitochondria. It is possible that the conformations in equilibrium may change according to the requirements of the mitochondrion. Because a mitochondrion is a primitive organelle, such a minute structural change may provide an economical mechanism for functional regulation of hmtRNA. However, once the T12311C mutation was introduced into the U48C mutant, the T-stem slip ‘switch’ was destroyed, preventing the change from the Ta alignment to the Tb alignment and inhibiting the regulation of the charging capacity. Thus, we hypothesize that it is either the fixed charging capacity or the frozen structure of the U48C mutant that may be the cause of the related disease.
Usually, in vitro transcription is an efficient approach to study the contribution of an individual nucleotide to tRNA structure and function (6,8,24–26). Owing to the absence of post-transcriptional modifications, different properties between the transcripts and their native counterparts were discovered (18,27,37,38). Based on this knowledge, although the hmtRNALeu(CUN) transcript appears to fold correctly and is aminoacyable, it mimics the native tRNA mainly at an immature level. Further in vivo study will be helpful to elucidate the mechanism of the related diseases.
Acknowledgments
This work was funded by the Natural Science Foundation of China (Grant 30270310 and 30330180), Committee of Science and Technology in Shanghai (Grant 02DJ140567) and the 863 projects of China (Grant 2004AA235091). Funding to pay the Open Access publication charges for this article was provided by the National Science Foundation of China (Grant 30270310).
Conflict of interest statement. None declared.
REFERENCES
- 1.Söll D. Transfer RNA: an RNA for all season. In: Gestelandm R.F., Atkins J.F., editors. The RNA World. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 157–183. [Google Scholar]
- 2.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Helm M., Brule H., Friede D., Giegé R., Putz D., Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulkes T., Hanna M.G. Human mitochondrial DNA diseases. Adv. Drug Deliv. Rev. 2001;49:27–43. doi: 10.1016/s0169-409x(01)00124-7. [DOI] [PubMed] [Google Scholar]
- 5.Florentz C., Sissler M. Disease-related versus polymorphic mutations in human mitochondrial tRNAs. Where is the difference? EMBO Rep. 2001;2:481–486. doi: 10.1093/embo-reports/kve111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelley S.O., Steinberg S.V., Schimmel P. Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nature Struct. Biol. 2000;7:862–865. doi: 10.1038/79612. [DOI] [PubMed] [Google Scholar]
- 7.Wittenhagen L.M., Kelley S.O. Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem. Sci. 2003;28:605–611. doi: 10.1016/j.tibs.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Levinger L., Morl M., Florentz C. Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res. 2004;32:5430–5441. doi: 10.1093/nar/gkh884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura Y., Gojobori T., Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hattori Y., Goto Y., Sakuta R., Nonaka I., Mizuna Y., Horai S. Point mutations in mitochondrial tRNA genes: sequence analysis of chronic progressive external ophthalmoplegia (CPEO) J. Neurol. Sci. 1994;125:50–55. doi: 10.1016/0022-510x(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 11.Dirheimer G., Keith G., Dumas P., Westhof E. Primary, secondary and tertiary structures of tRNAs. In: Söll D., RajBhandary U.L., editors. tRNA Structure, Biosynthesis and Function. Washington DC: American Society for Microbiology; 1995. pp. 93–126. [Google Scholar]
- 12.Li Y., Wang E.D., Wang Y.L. A modified procedure for fast purification of T7 RNA polymerase. Protein Expr. Purif. 1999;16:355–358. doi: 10.1006/prep.1999.1083. [DOI] [PubMed] [Google Scholar]
- 13.Yao Y.N., Wang L., Wu X.F., Wang E.D. Human mitochondrial leucyl-tRNA synthetase with high activity produced from Escherichia coli. Protein Expr. Purif. 2003;30:112–116. doi: 10.1016/s1046-5928(03)00097-4. [DOI] [PubMed] [Google Scholar]
- 14.Hao R., Yao Y.N., Zheng Y.G., Xu M.G., Wang E.D. Reduction of mitochondrial tRNALeu(UUR) aminoacylation by some MELAS-associated mutations. FEBS Lett. 2004;578:135–139. doi: 10.1016/j.febslet.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Puglisi J.D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silberklang M., Gillum A.M., RajBhandary U.L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- 18.Helm M., Brule H., Degoul F., Cepanec C., Leroux J.P., Giegé R., Florentz C. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998;26:1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peattie D.A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol A., Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- 21.Li T., Li Y., Guo N.N., Wang E.D., Wang Y.L. Discrimination of tRNALeu isoacceptors by the insertion mutant of Escherichia coli leucyl-tRNA synthetase. Biochemistry. 1999;38:9084–9088. doi: 10.1021/bi9901984. [DOI] [PubMed] [Google Scholar]
- 22.Xu M.G., Chen J.F., Martin F., Zhao M.W., Eriani G., Wang E.D. Leucyl-tRNA synthetase consisting of two subunits from hyperthermophilic bacteria Aquifex aeolicus. J. Biol. Chem. 2002;277:41590–41596. doi: 10.1074/jbc.M205126200. [DOI] [PubMed] [Google Scholar]
- 23.Chang Y.N., Pirtle I.L., Pirtle R.M. Nucleotide sequence and transcription of a human tRNA gene cluster with four genes. Gene. 1986;48:165–174. doi: 10.1016/0378-1119(86)90362-8. [DOI] [PubMed] [Google Scholar]
- 24.Sohm B., Frugier M., Brule H., Olszak K., Przykorska A., Florentz C. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 2003;328:995–1010. doi: 10.1016/s0022-2836(03)00373-5. [DOI] [PubMed] [Google Scholar]
- 25.Kelley S.O., Steinberg S.V., Schimmel P. Fragile T-stem in disease-associated human mitochondrial tRNA sensitizes structure to local and distant mutations. J. Biol. Chem. 2001;276:10607–10611. doi: 10.1074/jbc.M008320200. [DOI] [PubMed] [Google Scholar]
- 26.Roy M.D., Wittenhagen L.M., Vozzella B.E., Kelley S.O. Interdomain communication between weak structural elements within a disease-related human tRNA. Biochemistry. 2004;43:384–392. doi: 10.1021/bi035711z. [DOI] [PubMed] [Google Scholar]
- 27.Sohm B., Sissler M., Park H., King M.P., Florentz C. Recognition of human mitochondrial tRNALeu(UUR) by its cognate leucyl-tRNA synthetase. J. Mol. Biol. 2004;339:17–29. doi: 10.1016/j.jmb.2004.03.066. [DOI] [PubMed] [Google Scholar]
- 28.Larkin D., Williams A., Martinis S., Fox G. Identification of essential domains for Escherichia coli tRNALeu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res. 2002;30:2103–2113. doi: 10.1093/nar/30.10.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tocchini-Valentini G., Saks M.E., Abelson J. tRNA leucine identity and recognition sets. J. Mol. Biol. 2000;298:779–793. doi: 10.1006/jmbi.2000.3694. [DOI] [PubMed] [Google Scholar]
- 30.Asahara H., Nameki N., Hasegawa T. In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J. Mol. Biol. 1998;283:605–618. doi: 10.1006/jmbi.1998.2111. [DOI] [PubMed] [Google Scholar]
- 31.Soma A., Uchiyama K., Sakamoto T., Maeda M., Himeno H. Unique recognition style of tRNALeu by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol. 1999;293:1029–1038. doi: 10.1006/jmbi.1999.3219. [DOI] [PubMed] [Google Scholar]
- 32.Soma A., Kumagai R., Nishikawa K., Himeno H. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNALeu. J. Mol. Biol. 1996;263:707–714. doi: 10.1006/jmbi.1996.0610. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe Y., Tsurui H., Ueda T., Furushima R., Takamiya S., Kita K., Nishikawa K., Watanabe K. Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. J. Biol. Chem. 1994;269:22902–22906. [PubMed] [Google Scholar]
- 34.Ishitani R., Nureki O., Nameki N., Okada N., Nishimura S., Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 35.Randau L., Schauer S., Ambrogelly A., Salazar J.C., Moser J., Sekine S., Yokoyama S., Söll D., Jahn D. tRNA recognition by glutamyl-tRNA reductase. J. Biol. Chem. 2004;279:34931–34937. doi: 10.1074/jbc.M401529200. [DOI] [PubMed] [Google Scholar]
- 36.Hauenstein S., Zhang C.M., Hou Y.M., Perona J.J. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nature Struct. Mol. Biol. 2004;11:1134–1141. doi: 10.1038/nsmb849. [DOI] [PubMed] [Google Scholar]
- 37.Kirino Y., Yasukawa T., Ohta S., Akira S., Ishihara K., Watanabe K., Suzuki T. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl Acad. Sci. USA. 2004;101:15070–15075. doi: 10.1073/pnas.0405173101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sissler M., Helm M., Frugier M., Giegé R., Florentz C. Aminoacylation properties of pathology-related human mitochondrial tRNA(Lys) variants. RNA. 2004;10:841–853. doi: 10.1261/rna.5267604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]