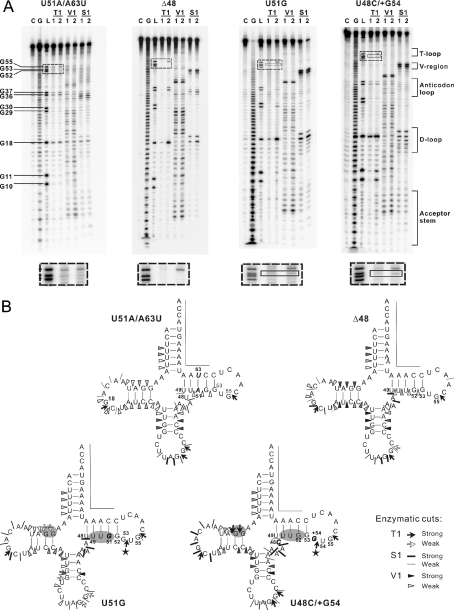
Figure 4.
Enzymatic probing of the representative tRNALeu(CUN) variants. (A) Cleavage products of 5′-labeled molecules after treatment with nucleases, displayed on autoradiograms of 12% polyacrylamide gels. C stands for H2O ladder; G for G ladder; L for alkaline ladder; T1 for RNase T1 digestion ladder; V1 for RNase V1 digestion ladder; S1 for nuclease S1 digestion ladder; numbers 1 and 2 refer to increasing concentrations of nuclease. The RNase T1 cleavage product denoted in the solid-line square indicates the enlarged T-loop. The band difference can be easily seen in the dashed-line squares. (B) Location of enzymatic cleavage sites on cloverleaf diagrams of the transcripts. Specifications and intensities of cuts are as indicated in the key. Nucleotides that could not be tested because of technical limitations are marked by a line. Mutated nucleotides are shown in italics. Regions with different cleavage pattern are highlighted by gray background. The pentacles denote the RNase T1 cleavage specific for the enlarged T-loop.