Figure 4.
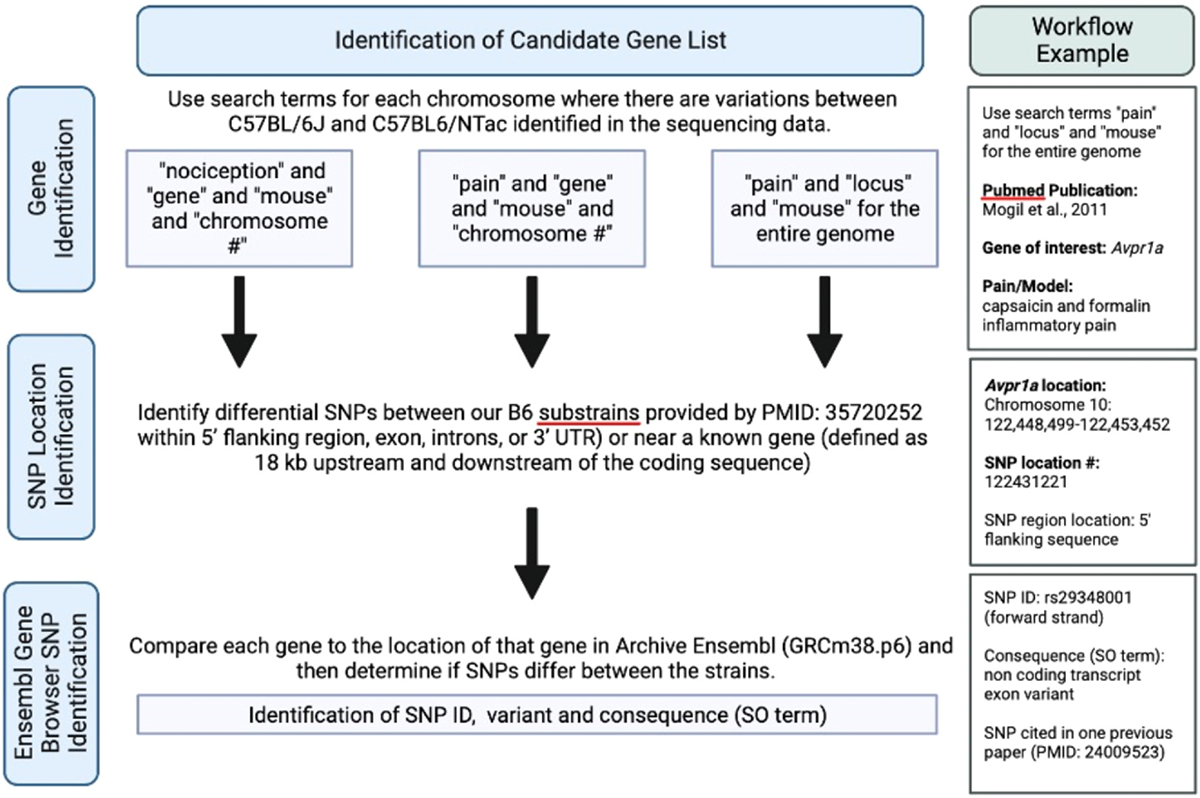
Workflow for identifying pain candidate genes from whole-genome sequencing data. In brief, we performed a systematic review and collected a list of genes related to visceral pain, nociception, and/or pain assays. From this list, we used whole-genome sequencing data published in Mortazavi et al21 to compare the location of our genes of interest to locations of possible differential SNPs between BL/6J and BL/6NTac. It is important to note Archive Ensembl was used to match and compare whole-genome sequencing data to the appropriate gene loci. To the right of the general flow is an example workflow used to identify an intergenic SNP within the 5′ region of Avpr1a.
