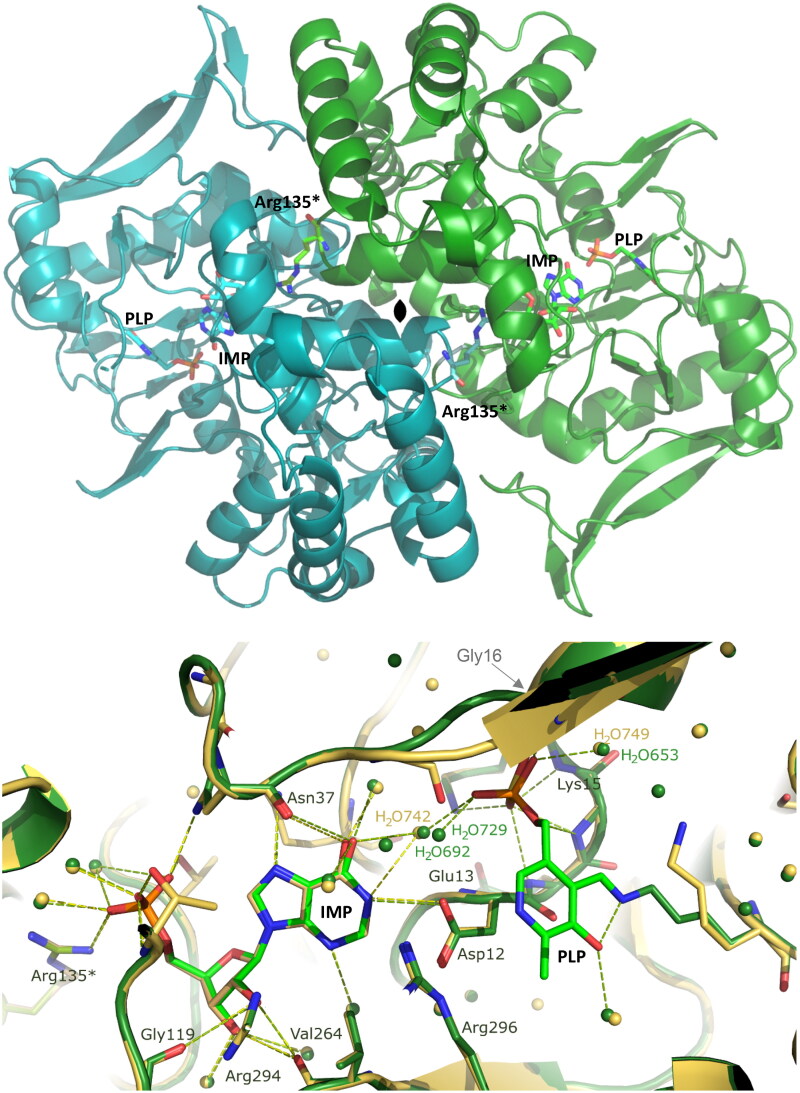
Figure 5.
Upper panel: The overall structure of the H. pylori AdSS dimer, which is formed by two identical monomers (shown in green and turquoise) as the result of a two-fold symmetry axis (denoted in the centre). The active sites are located on the opposite sides of the monomer-monomer interface and have the form of elongated clefts lying near the surface of the protein. The side-chain of amino acid Arg135 from each monomer (marked with an asterisk) completes the active site of the neighbouring subunit. The C-terminus in each monomer, to which the histidine tag is attached but not visible in the electron density, is positioned on the opposite side of the protein compared to the active site. Lower panel: Active site of the AdSS ternary complex with PLP and IMP (green, 8QWA) overlaid with the active site of the binary complex of the enzyme with only IMP (yellow, 7PVO) 14. Hydrogen bond network observed in both complexes is shown with dashed lines, green and yellow, respectively. Water molecules are represented by green and yellow spheres (the numbers of some of them are shown and correspond to their numbers in the respective PDB files). Gly16, which is here covered by the beta sheet of the structure, and which interacts through the main chain N atom via a hydrogen bond with the phosphate group of PLP (see Table 3), is shown by the arrow. Arg135 from the adjacent subunit (marked with an asterisk) is seen to be involved in IMP binding at the active site located in the neighbouring monomer of the dimer.