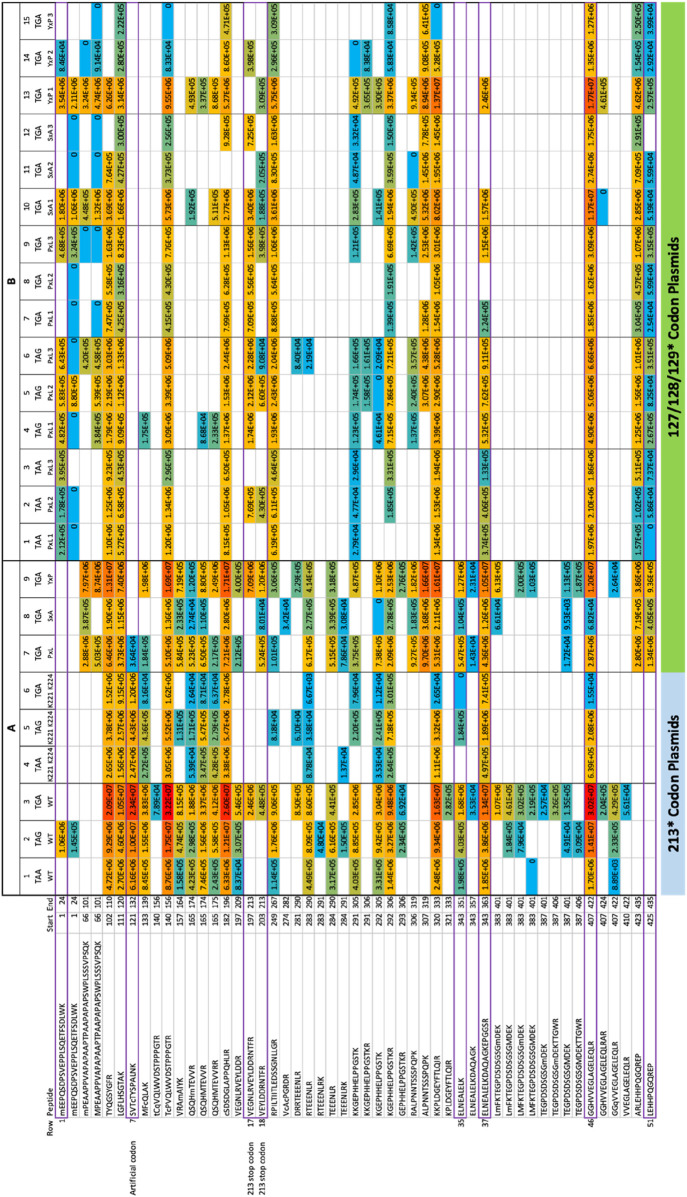
FIGURE 3.
Overall summary of peptide identification that corresponds to the wild-type p53 sequence over all the replicates. (A) The transient transfection methodology. We initially transiently transfected three different plasmids containing the three different nonsense termination codons at codon 213 of the p53 gene (columns 1–3); peptides matching the wild-type p53 sequence using PEAKS that were acquired from the 213-TAA plasmid are presented in Column 1; the 213-TAG plasmid expression peptides are Column 2; and the 213-TGA plasmid expressing peptides are in Column 3. Overall, 51 different overlapping tryptic and semi-tryptic peptides were detected that match the wild-type p53 sequence in all three samples (see Figure 2A as a representative summary of overall peptide sequence coverage in the p53 gene and a detailed summary of specific peptides in columns 1–3, row labelled 1 at the top and row 51 at the bottom). A second independent transient transfection of the three different p53 expression plasmids was also performed (columns 4–6) containing the three different termination codons of p53 at codon 213 except these also contained two E-K mutations (221-EPPE-224; 221-KPPK-224) in order to introduce tryptic cleavage sites near the 213 termination codon in attempts to recover more tryptic peptides from this region; peptides matching the wild-type p53 sequence using PEAKS that were acquired from the 213-TAA/221-KPPK-224 plasmid are presented in Column 4; the 213-TAG/221-KPPK-224 plasmid expression peptides are Column 5; and the 213-TGA/221-KPPK-224 plasmid expressing peptides are in Column 6. A third experiment involved transient transfection of three different plasmids with the same TGA mutation at codons 127, 128, and 129 (Figure 3A, columns 7–9). (B) The stably integrated p53 gene methodology. A fourth independent experimental set was performed in which, after transfection, cells were selected with new sets of plasmids containing a hygromycin resistance gene for 4 weeks in the presence of Hygromycin B to maintain in culture only those cells that are carrying the integrated, tagged p53 with the nonsense mutations (Figure 3B). The overall wild-type p53 tryptic peptide recovery was very similar (Figures 3A vs. Figure 3B), although more peptides were recovered in the transient transfection vs. the stably integrated plasmid (Figures 3A vs. Figure 3B; rows 1–51). This might be due to the fact that transient transfection can produce an artificially high level of p53 protein, whereas stable cells which integrated the plasmid might be selected for the production of lower amounts of p53. Finally, in addition to the use of PEAKS to identify and quantify peptides, we also used Proteome Discoverer™ to quantify peptides (Supplementary Figure S5), with generally consistent results compared to PEAKS (Supplementary Figures S2–S4; Figure 2B). These data have: (i) the codon 129 (PxL) expression plasmids in triplicates (columns 1–3, TAA; columns 4–6, TAG; and columns 7–9, TGA); (ii) the codon 128 (SxA) expression plasmids in triplicate (columns 10–12, TGA); (iii) the codon 127 (YxP) expression plasmids in triplicate (columns 13–15, TGA). The numbers in each box represent ion intensity.