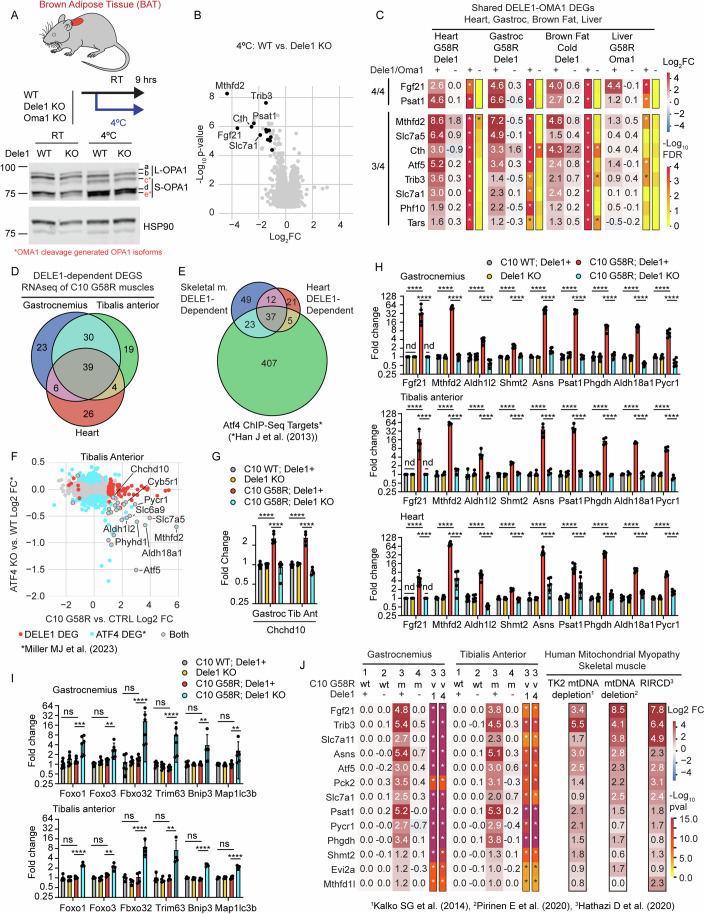
Figure 6. The DELE1 mt-ISR is observed in several mitochondria-rich tissues and has greatest overlap between heart and skeletal muscle.
(A) Dele1 KO and WT littermates were challenged with cold stress for 9 h and interscapular BAT was analyzed by immunoblotting for OPA1 cleavage by OMA1. OMA1 cleavage generated OPA1 isoforms are in indicated in red as c* and e*. (B) Volcano plot of global gene expression changes in cold stressed Dele1 KO vs. WT littermates measured by microarray; significant genes (FDR < 0.05 and |Log2FC| > 1) are in black. Statistics were performed as described in Methods for all microarray-based transcriptomics. N = 7 mice per group. (C) Heat map shows overlap among DELE1-dependent DEGS in four mitochondrial-rich tissues: heart, gastrocnemius (gastroc) skeletal muscle, and liver from C10 G58R mice, in addition to BAT from mice subjected to cold stress. BAT microarray data from same dataset that also appears in Appendix Fig. S1B. C10 G58R heart microarray data are from the same dataset that also appears in Appendix Fig. S1B, Fig. 2E, and Fig. 4B. Statistics on all microarray data (which also appear in Dataset EV6) were performed as described in Methods for all microarray-based transcriptomics. FDR values were corrected for multiple comparisons across all transcripts in dataset. N = 4 for all groups except for C10 G58R; Dele1+ liver, which was N = 3. (D) Venn diagram shows intersection of C10 G58R DELE1-dependent DEGs from three muscles (gastrocnemius, tibalis anterior, and heart), measured by RNA-Seq at P28 (data also available in Dataset EV7–EV9). (E) Venn diagram shows intersection of skeletal and and heart muscle DELE1-dependent DEGs as in (D) with ATF4 target genes, from previously published ATF4 ChIP-Seq dataset (Han et al, 2013). (F) Scatterplot shows correlation between genes that increase in tibalis anterior of P28 C10 G58R mice vs. CTRL and those that decrease in abundance in tibalis anterior from 6-month-old ATF4 skeletal muscle knockout mice vs. CTRL, from a previously published dataset (Miller et al, 2023). (G, H) Individual data is shown for select DELE1-dependent genes detected from skeletal muscle and/or heart from P28 C10 G58R mice, by RNA-Seq (data for all detected genes in Dataset EV7–EV9). Fgf21 was below detection limit (not detected, “nd”) in all but the C10 G58R; Dele1+ condition. Significance was first tested with a two-way ANCOVA including genotype and sex as variables. For post hoc testing) after ANCOVA (shown in Figure), the general linear hypotheses was used in conjunction with the multiple comparisons of means (“mcp”) function to test for all possible two-way comparisons via “Tukey” method. For (G), p-values were 1.98E−08 and 6.98E−07 (for bottom row) and 7.12E−10 and 6.62E−08 (for top row). For (H, top graph, gastrocnemius) p-values for C10 WT; Dele1+ vs. C10 G58R; Dele1+ (for genes left to right) were 0, 0, 2.58E−09, 1.89E−15, 0, 0, 0, 0, and 1.11E−16, and for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO (for genes left to right) were 0, 0, 1.19E−12, 2.50E−13, 0, 0, 0, 0, and 0. For (H, middle graph, tibialis anterior) p-values for C10 WT; Dele1+ vs. C10 G58R; Dele1+ (for genes left to right) were 4.37E−07, 0, 4.63E−06, 7.99E−09, 4.97E−11, 0, 0, 2.33E−15, 3.42E−13 and for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO (for genes left to right) were 8.51E−08, 0, 2.24E−05, 1.37E−08, 1.24E−12, 0, 2.22E−16, 7.77E−16, and 0. For (H, top graph, heart) p-values for C10 WT; Dele1+ vs. C10 G58R; Dele1+ (for genes left to right) were 1.57E−10, 0, 1.42E−11, 1.63E−12, 0, 2.71E−09, 0, 0, and 0, and for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO (for genes left to right) were 6.21E−10, 1.22E−12, 1.55E−15, 3.22E−15, 1.16E−13, 0.0001011, 0, 0, 0. In all graphs, **** indicates p ≤ 0.0001, respectively, and “ns” not significant. Error bars represent SD. N ≥ 4 mice per group (genotype). (I) individual data is shown for select genes significantly elevated in skeletal muscles of only C10 G58R; Dele1 KO and no other genotypes relative to control. Significance was first tested with a two-way ANCOVA including genotype and sex as variables. For post hoc testing) after ANCOVA (shown in Figure), the general linear hypotheses was used in conjunction with the multiple comparisons of means (“mcp”) function to test for all possible two-way comparisons via “Tukey” method. For gastrocnemius (top graph), p-values for C10 WT; Dele1+ vs. C10 G58R; Dele1+ (for genes left to right) were 0.7386, 0.3720, 0.8302, 0.8152, 0.9998, and 0.9999, and for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO (for genes left to right) were 0.0007080, 0.001020, 7.28E−05, 2.80E−05, 0.003794, 0.005845. For tibialis anterior (bottom graph), p-values for C10 WT; Dele1+ vs. C10 G58R; Dele1+ (for genes left to right) were 0.9919, 0.3088, 0.9999, 0.9984, 0.2705, 0.9920, and for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO (for genes left to right) were 3.52E−05, 0.002405, 1.66E−05, 0.001349, 1.16E−07, 1.30E−08. In all graphs, **, ***, **** indicates p ≤ 0.01, 0.001, 0.0001, respectively, and “ns” not significant. Error bars represent SD. N ≥ 4 mice per group (genotype). (J) Heat map of 13 mouse genes that are DELE1-dependent in C10 G58R gastrocnemius and tibialis anterior model (as in D) and have human orthologs that are upregulated by >2-fold (on average) in three previously published datasets from mitochondrial myopathy patients (Hathazi et al, 2020; Pirinen et al, 2020; Kalko et al, 2014). P-values were corrected for multiple comparisons across all transcripts in dataset. Source data are available online for this figure.