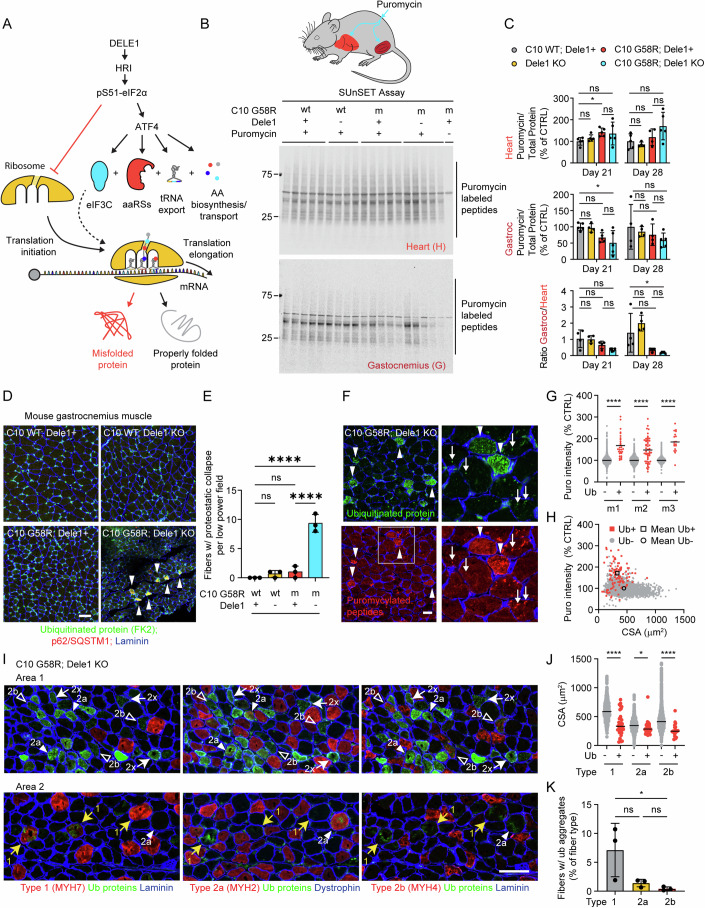
Figure 7. DELE1 mt-ISR promotes translation-associated proteostasis in striated muscle.
(A) Model depicts predicted effects of the DELE1 mt-ISR on protein translation including acute inhibition of translation initiation by pS51-eIF2α, resumption of protein initiation following the transcriptional upregulation of Eif3c, and facilitation of translation elongation through increased production of protein synthesis intermediates by the coordinated upregulation of (1) genes related amino biosynthesis and transport, (2) Xpot to mediate tRNA export from the nucleus to the cytosol, and (3) aminoacyl-tRNA synthases (aaRSs) to promote aminoacyl conjugation to tRNAs. Protein translation dynamics and fidelity can affect the proportion of newly translated proteins that fold properly vs. misfold. (B, C) Measurement of in vivo protein synthesis in C10 G58R; Dele1 KO mice and indicated littermates at P21 and P28, using the SUnSET assay. Blot for P21 timepoint is shown in (B). Mice were injected with the aminoacyl-tRNA ortholog, puromycin, sacrificed 30 min later, and puromycin incorporation into newly synthesized polypeptides in heart and gastrocnemius tissue lysates was measured by immunoblotting. Statistical analysis performed using Welch ANOVA and Dunnett’s T3 multiple comparisons test. (Top graph, heart, Day 21) adjusted p-values were 0.4894 for C10 WT; Dele1+ vs. Dele1 KO, 0.0236 for C10 WT; Dele1+ vs. C10 G58R; Dele1+, 0.5341 for C10 WT; Dele1+ vs. C10 G58R; Dele1 KO, and 0.9978 for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO. (Top graph, heart, Day 28) adjusted p-values were 0.8419 for C10 WT; Dele1+ vs. Dele1 KO, 0.8923 for C10 WT; Dele1+ vs. C10 G58R; Dele1+, 0.2250 for C10 WT; Dele1+ vs. C10 G58R; Dele1 KO, and 0.4979 for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO. (Middle graph, gastroc, Day 21) adjusted p-values were 0.9990 for C10 WT; Dele1+ vs. Dele1 KO, 0.1338 for C10 WT; Dele1+ vs. C10 G58R; Dele1+, 0.0136 for C10 WT; Dele1+ vs. C10 G58R; Dele1 KO, and 0.7086 for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO. (Middle graph, gastroc, Day 28) adjusted p-values were 0.9841 for C10 WT; Dele1+ vs. Dele1 KO, 0.9399 for C10 WT; Dele1+ vs. C10 G58R; Dele1+, 0.7107 for C10 WT; Dele1+ vs. C10 G58R; Dele1 KO, and 0.8419 for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO. (Top graph, gastroc/heart ratio, Day 21) adjusted p-values were 0.9999 for C10 WT; Dele1+ vs. Dele1 KO, 0.6361 for C10 WT; Dele1+ vs. C10 G58R; Dele1+, 0.2390 for C10 WT; Dele1+ vs. C10 G58R; Dele1 KO, and 0.1941 for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO. (Top graph, gastroc/heart ratio, Day 28) adjusted p-values were 0.8820 for C10 WT; Dele1+ vs. Dele1 KO, 0.4304 for C10 WT; Dele1+ vs. C10 G58R; Dele1 +, 0.3415 for C10 WT; Dele1+ vs. C10 G58R; Dele1 KO, and 0.1941 for C10 G58R; Dele1+ vs. C10 G58R; Dele1 KO. In all graphs, * indicates p ≤ 0.05 and “ns” not significant. N ≥ 4 mice per group (genotype). Error bars represent SD. (D) Representative immunofluorescence images of gastrocnemius muscle from P28 C10 G58R; Dele1 KO mice and indicated littermates, showing fibers with many or confluent aggregates of ubiquitinated proteins co-localized with the aggregate-forming adapter protein p62, suggesting proteostatic collapse (arrowheads) in low power (20×) images (top panels). Scale bars = 50 μm. (E) Quantification of (D). N = 3 mice per genotype with 30 low power fields counted per genotype. Low-power field size was 397.75 μm × 397.75 μm. Statistical analysis performed using ordinary one-way ANOVA with Šidák’s multiple comparisons test, with a single pooled variance. N = 3 in each group. P = 0.8857 for C10 WT; Dele1+ vs. C10 WT; Dele1 KO. P = 0.6605 for C10 WT; Dele1+ vs. C10 G58R; Dele1 +. ns = non-significant. **** =< 0.0001. Error bars represent SD. (F) Representative immunofluorescence images of gastrocnemius muscle from P28 C10 G58R; Dele1 KO mice injected with puromycin 30 min prior to sacrifice as in (B). Muscle cross-sections were immunostained for ubiquitinated proteins (using the FK2 antibody) (green), puromycin (red), and laminin (blue). Arrowheads indicate muscle fibers containing many or confluent aggregates of ubiquitinated proteins that were also co-stained for elevated puromycylated polypeptides. Arrows indicate individual aggregates of ubiquitin proteins that also contain puromycylated polypeptides. Scale bars = 20 μm. (G, H) Quantification of (F). Individual myofibers were automatically segmented using the Laminin immunofluorescence to define the myofiber border. Average puromycin fluorescence intensity and cross-sectional area (CSA) were measured for each myofiber. Separately myofibers were manually scored as containing many or confluent aggregates of ubiquitinated proteins (Ub+ or Ub- muscle fibers, respectively). The average puromycin intensity for Ub+ and Ub- myofiber is shown in graph separately for three mice (m1–3). N = 3 mice with 10 high power fields counted per mouse. The scatterplot in (H) shows the relationship between myofiber CSA and puromycin intensity for all myofibers analyzed in (G). Fibers with CSA < 50 μm were excluded from analysis. Statistical analysis was performed using Kruskal–Wallis test with Dunn’s multiple comparisons test as data was non-parametrically distributed for (G). (I) Representative confocal images show three consecutive sections through two areas of gastrocnemius from C10 G58R; Dele1 KO mice triple stained for muscle fiber type 1, 2a, or 2b (red) and FK2 to detect ubiquitin protein aggregates (green) and a sarcolemma marker protein laminin or dystrophin (blue). Fiber type 2x is defined by the absence of any other fiber marker in the consecutive sections. Areas 1 and 2 are magnifications of the boxed areas in Appendix Fig. S16, which shows the whole tissue section. Scale bar = 100 μm. Yellow arrow = type 1 fiber, solid white arrowhead = type 2a fiber, open white arrowhead = 2b fiber, and solid white arrow = 2x fiber. (J) Graph quantifying the CSA of fibers with and without aggregates of ubiquitinated proteins, separated by fiber type as in (I). Gastrocnemius from three C10 G58R; Dele1 KO mice were analyzed. Statistical analysis was performed using Kruskal–Wallis test with Dunn’s multiple comparisons test as data was non-parametrically distributed. (K) Graph quantifying the proportion of each fiber types that contained aggregates of ubiquitinated proteins from tissue stained as in (I). Gastrocnemius from three C10 G58R; Dele1 KO mice were analyzed. Statistical analysis was performed using Kruskal–Wallis test with Dunn’s multiple comparisons test as data was non-parametrically distributed. N = 3 in each group. P = 0.4081 for percent of type 1 myofibers with FK2 aggregates vs. percent of type 2a myofibers with FK2 aggregates. P = 0.0338 for percent of type 1 myofibers with FK2 aggregates vs. percent of type 2b myofibers with FK2 aggregates. P = 0.8902 for percent of type 1 myofibers with FK2 aggregates vs. percent of type 2b myofibers with FK2 aggregates. ns = non-significant. * = <0.05. Error bars represent SD. Source data are available online for this figure.