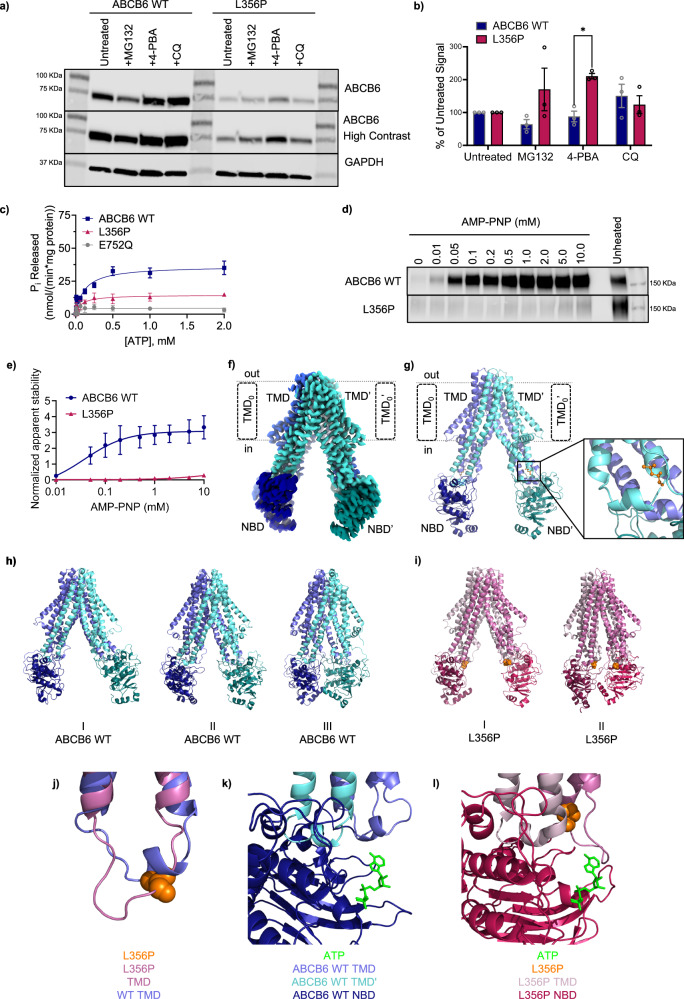
Fig. 2. Characterization of the L356P mutation.
a Western blot shows L356P is disproportionately stabilized by 4-phenylbutyrate (4-PBA) compared to ABCB6 WT. MG132 and chloroquine (CQ) also show a less significant effect on expression (representative data shown). b Band signals from (a) and replicates were quantitated and the percentage of untreated signal was calculated (see Supplementary Methods). ABCB6 WT is shown in indigo, L356P is shown in magenta. Analysis using an unpaired two-tailed Welch’s T-test and found no significant differences between ABCB6 WT and L356P except for with 4-PBA treatment (p = 0.00262, N = 3 biological replicates per treatment.) Data are reported as mean ± SEM. c L356P (magenta) has significantly decreased ATPase activity compared to ABCB6 WT (indigo). The catalytically inactive E752Q mutant is shown in grey. Data are reported as mean ± SEM, N = 15 experiments for ABCB6 WT from five biological replicates, N = 8 experiments for E752Q from three biological replicates, and N = 4 for L356P from two biological replicates. d Western blot of isothermal shift assay shows L356P exhibits little thermal stabilization with AMP-PNP treatment, suggesting L356P cannot interact with ATP (representative data shown). e Band signals from (d) and replicates were quantitated. Data are reported as mean ± SEM. N = 3 technical replicates for ABCB6 WT (indigo) and L356P (magenta). f Cryo-EM structure of ABCB6 WT (EMDB ID: EMD-46724, PDB ID: 9DBQ) and resulting cartoon model (g). Nucleotide binding domains (NBDs) are shown in darker tones while transmembrane domains (TMDs) are colored in lighter tones. L356 (orange) is located near the coupling helix of ABCB6 (inset). Predominant conformations of ABCB6 WT (h) and L356P (i) over the combined 1500 ns of MD simulation as selected from hierarchical agglomerative clustering analysis (j) Overlay of ABCB6 WT (purple) and L356P (pink) showing the change in conformation of the coupling helix. A single chain of the homodimer has been shown for clarity, however similar changes are observed in both monomers. Overlay of ATP from ABCB10 (PDB ID: 4AYT) onto ABCB6 WT (k) or L356P (l) showing the change in conformation of the ATP binding site. Source data are provided in the Source Data file.