FIGURE 1.
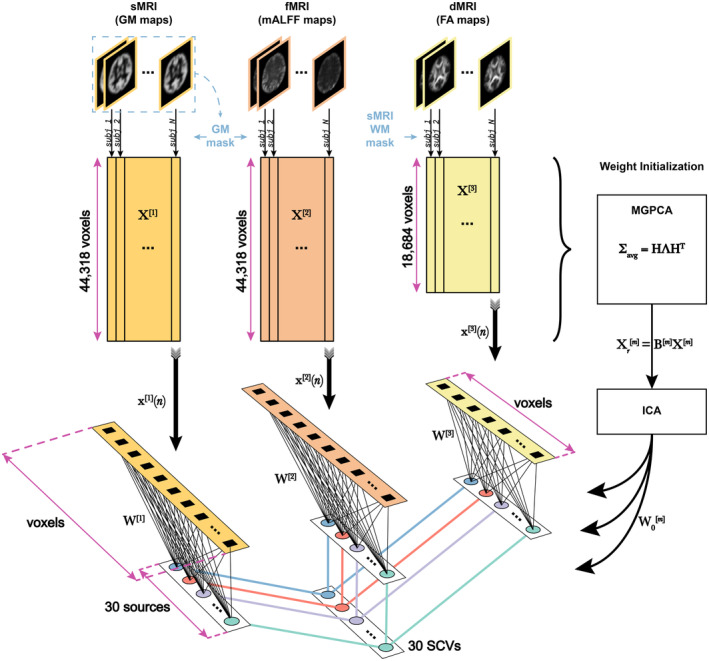
Multimodal IVA (MMIVA) fusion. First, we opted for cross‐modality co‐registration and masking to select the features used in this 3‐way fusion analysis. Second, we pursued a multimodal group PCA (MGPCA) preprocessing strategy that accounts for the total variance of each modality, ensuring they contribute equally to the estimation of common directions. Third, the proposed MGPCA+ICA initialization of the unmixing weights serves to “pre‐align” the latent subspaces. Nevertheless, learning of the final unmixing is still done with the full 30‐by‐voxels matrices, that is, MMIVA leverages the full dimensionality of the spatial features (no data reduction), allowing for full interaction between modalities. Fourth, the choice of Kotz parameters (, , ) is such that it addresses potential limitations of the zero‐mean Laplace distribution typically employed in IVA literature, namely that its derivative at 0 is not well‐defined (discontinuous), thus eliminating certain risks for numerical instability. Fifth, unlike typical IVA methods, the choice of Kotz distribution is sensitive to all‐order statistics, not just second‐order (also known as linear dependence). Sixth, sources are expression levels, meaning that the patterns in the expression levels over subjects (not the spatial maps) are statistically independent of one another and linked across modalities. This is a sensible choice because it treats subjects as the observations, which is atypical of blind source separation in neuroimaging due to the often low sample sizes (i.e., sample‐poor regimes, low ). Finally, MMIVA is the first mature application of MISA on real multimodal datasets (although a preliminary version that did not account for site effects and did not include the study on patients appeared at the IEEE EMBC 2021 conference (Damaraju et al. 2021)).
