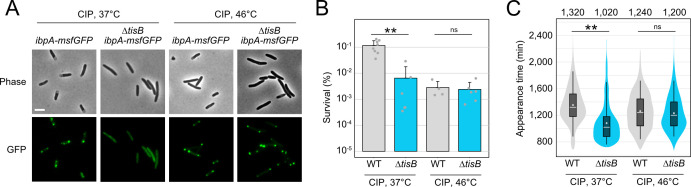
Fig 8.
Heat-induced protein aggregates affect recovery from CIP. (A) Strain MG1655 ibpA-msfGFP and ΔtisB ibpA-msfGFP were treated with CIP (10 µg/mL; 1,000× MIC) during the exponential phase (OD600 ~0.4) for 6 hours at 37°C or 46°C. Phase contrast images are displayed together with corresponding fluorescence images (GFP). White bars represent a length scale of 2 µm. (B) WT MG1655 and a tisB deletion mutant were treated with ciprofloxacin (10 µg/mL; 1,000× MIC) during the exponential phase (OD600 ~0.4) for 6 hours at 37°C or 46°C. Pre- and post-treatment samples were used to determine relative CFU (%). Bars represent the mean of at least four biological replicates and error bars indicate the standard deviation. Dots show individual data points (WT 37°C: n = 8; ΔtisB 37°C: n = 6; WT 46°C: n = 4; ΔtisB 46°C: n = 6). ANOVA with post-hoc Tukey HSD was performed (**P < 0.01, ns: not significant). (C) WT MG1655 and a tisB deletion mutant were treated with ciprofloxacin (10 µg/mL; 1,000×MIC) during exponential phase (OD600 ~0.4) for 6 hours at 37°C or 46°C. ScanLag was applied to determine the colony appearance time after CIP treatment. Colony appearance times are illustrated as violin box plots. Colonies from at least three biological replicates were combined (WT 37°C: n = 1,431; ΔtisB 37°C: n = 272; WT 46°C: n = 476; ΔtisB 46°C: n = 1,026). The white dot indicates the mean. The respective median appearance time (white bar) is shown on top of each plot. The ΔtisB mutant was compared to the corresponding wild type MG1655 using a pairwise Wilcoxon rank-sum test (**P < 0.0001, ns: not significant).