Figure 4.
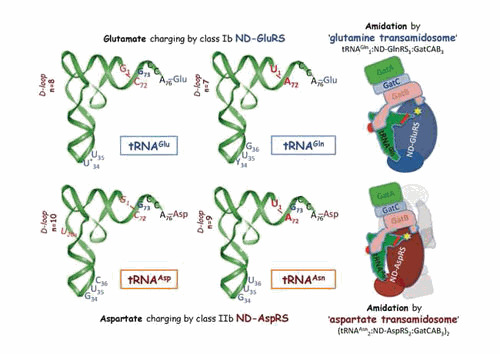
Dual tRNA aminoacylation by ND-GluRS and ND-AspRS and tRNA-dependent amino acid amidation, the two steps of the indirect pathway of glutaminyl-tRNAGln and asparaginyl-tRNAAsn formation. (Left) The two tRNA couples (tRNAGlu / tRNAGln and tRNAAsp / tRNAAsn) aminoacylated by ND-GluRS and ND-AspRS, respectively, and (Right) schematized representations of the T. maritima glutamine (3al0) (186) and T. thermophilus aspartate (3kfu) (187) transamidosomes. The main structural and functional features important for tRNA aminoacylation by ND-aaRSs and for tRNA-dependent conversion of the glutamyl and aspartyl residues into glutamine and asparagine are shown, as well as the amidation site (yellow star) in the transamidosomes. The major identity determinants for aminoacylation of the four tRNAs by their cognate aaRSs are shown in blue (6, 188). In Bacteria, position 34 is a modified U in tRNAGlu and a pyrimidine (Y) in tRNAGln. Notice the quite similar identity sets in the tRNAGlu / tRNAGln and tRNAAsp / tRNAAsn couples, in agreement with their dual aminoacylation by the ND-aaRSs. In bold red: U1–A72, the major identity determinants for amidation in tRNAGln and tRNAAsn; in red italics: notably the antideterminant G1–C72 pair that prevents glutamate and aspartate amidation in charged tRNAGlu and tRNAAsp (186, 189). The longer length of the D-loop in tRNAGlu and tRNAAsp (as compared to tRNAGln and tRNAAsn, a feature conserved in Bacteria [19]) is a further antideterminant that prevents amidation. Transamidosomes show an overall conserved organization based on the association of ND-aaRS, tRNA, and heterotrimeric GatCAB. Notice the Yqey domain of GatB that contacts the D-loop of tRNA and thereby plays a key role in transamidation. Notice further the different sizes of the two transamidosomes. While the glutamine transamidosome is formed by five entities (as seen in the figure), the much larger aspartate transamidosome is formed by 14 macromolecular entities (for clarity, only half of the structure is shown, with the second subunit of AspRS and its tRNA ligand shown in light grey). This architectural variation is due to structural differences in ND-GluRSs (class Ib monomers) and ND-AspRSs (class IIb dimers) and the correlated mechanistic differences in the aminoacylation and transamidation steps occurring within the two types of transamidosome (see Fig. 6 in “Aminoacylation of tRNA” and “Indirect pathways of specific tRNA aminoacylation for ribosome-mediated translation,” below, for details).
