Figure 22.
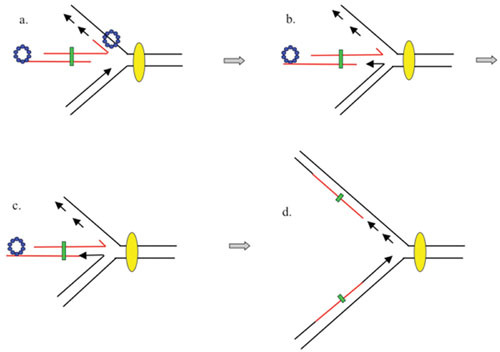
Proposed model for integration of dsDNA substrate with limited nonhomology into a replication fork. (A) Red-processed dsDNA end (Beta bound to a ssDNA overhang generated by Exo) invades a replication fork and promotes annealing to the lagging strand template. (B) The leading strand switches to the incoming duplex. Stalling and reversal of the fork generates a “chicken-foot” structure. (C) The leading strand polymerase fills in the gap previously generated by λ Exo, while DNA ligase connects the strands. (D) The Holliday junction branch migrates to the right and is reabsorbed, reestablishing a replication fork framework. The mutant base(s) form heteroduplexes (green boxes). Model taken from Court et al. (381).
