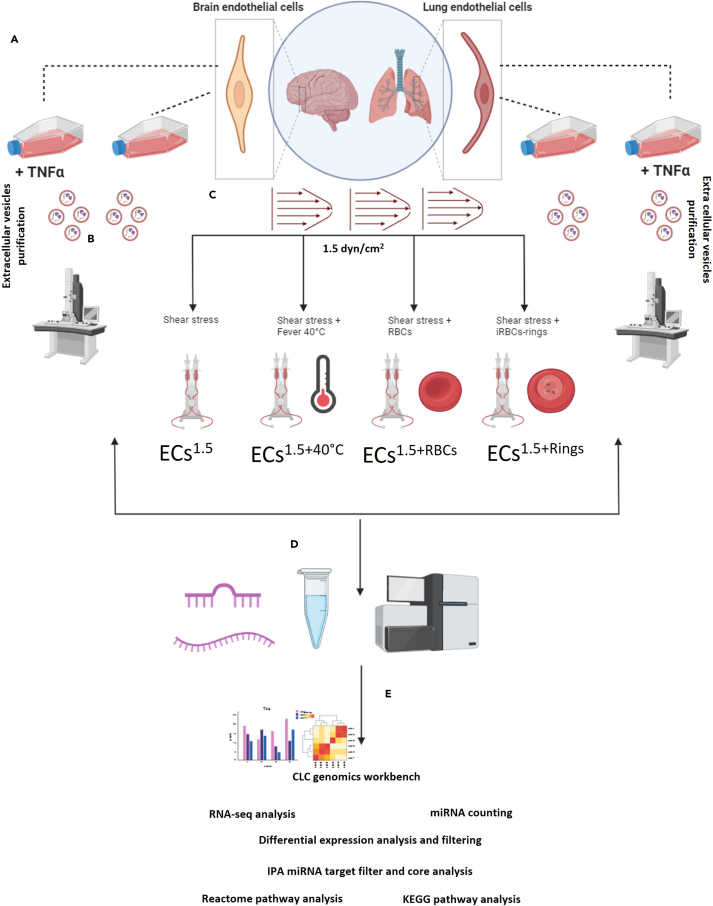
Figure 1.
Schematic overview of study workflow
(A) Primary brain endothelial cells (ECs) were cultured under static conditions without TNFα (HBMECstatic) or with TNFα (HBMECstatic+TNF) as were primary lung ECs (HMVEC-Lstatic or HMVEC-Lstatic+TNF). Culture supernatants containing EVs were harvested and sequentially centrifuged to isolate EVs.
(B) Transmission electron microscopy was performed to confirm the presence of EVs after purification.
(C) A laminar flow system was used to apply shear stress of 1.5 dyne/cm2. Stimuli were then added to the fluidic unit: (i) incubation at 40°C (ECs1.5+40°C), (ii) non-infected red blood cells (RBCs) (ECs1.5+RBCs), and (iii) infected RBCs (iRBCs) (ECs1.5+Rings).
(D) mRNA/miRNAs were then isolated from ECs and purified EVs and subjected to NGS sequencing.
(E) Raw data were extracted and subjected to RNA-seq analysis and miRNA counting using a CLC genomics workbench. Subsequently, differential expression analysis was performed. Finally, miRNA target filtration was performed followed by pathway analysis using Reactome and KEGG pathway analysis.