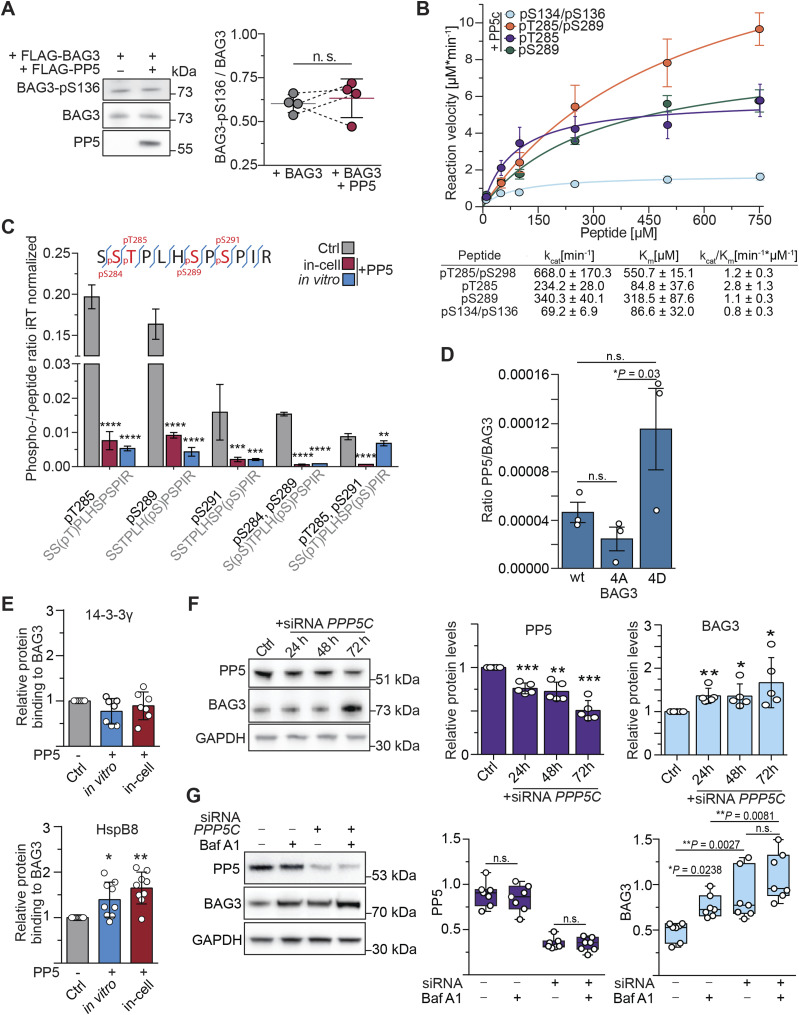
Figure 5. PP5 dephosphorylates a p-site cluster in BAG3 and enables HspB8 binding.
(A) Phosphorylation levels of BAG3-pS136 were compared in HEK293 cells overexpressing FLAG-BAG3 or both FLAG-BAG3 and FLAG-PP5 for 24 h. Immunoblot quantification demonstrates that PP5 overexpression does not affect BAG3-pS136 phosphorylation. Statistical significance was determined through t test (n = 4). (B) Michaelis–Menten kinetic parameters of PP5c were determined against the indicated mono- or bisphosphorylated peptides. Error bars represent the SEM of three independent repeats with technical duplicates. kcat/Km was calculated by comparison with a phosphate standard curve. (C) HEK293 cells overexpressed FLAG-BAG3 and were treated with 1 μM recombinant PP5 after lysis for 1 h (in vitro), or overexpressed FLAG-BAG3 and FLAG-PP5 simultaneously for 24 h (in-cell), or were not treated with phosphatase (Ctrl). FLAG-BAG3 was subjected to affinity enrichment and subsequently evaluated using LC-MS/MS. The theoretical phosphopeptide generated through trypsinization is shown at the top. Targeted phosphoproteomics quantification reveals the complete dephosphorylation of the p-site cluster for both PP5 treatments. Statistical significance was determined through two-way ANOVA (n = 4), error bars represent the SD (****P < 0.0001, ***P = 0.0002, *P = 0.0021). Indexed retention time (iRT). (D) FLAG-BAG3 mutants mimicking the phosphorylated (4D) and dephosphorylated (4A) state of the BAG3 p-site cluster were overexpressed in HEK293 cells followed by co-IP and MS analysis to assess PP5 binding to each variant compared with WT. Bars represent the mean (n = 3), with significance and error bars calculated using t test (*P = 0.03). (C, E) Samples were subjected to the same treatment as described for (C) to induce dephosphorylation of p-site cluster. This was followed by the analysis of protein quantities bound to BAG3 via immunoblotting, normalized to the amounts bound to the control sample. Statistical significance of changes in protein levels was assessed using data from seven or nine independent replicates, with means presented as bar plots. P-values were calculated using a one-way ANOVA with a 95% confidence interval and analyzed using PRISM/GraphPad version 6 (HspB8: *P = 0.0188; **P = 0.0002; 14-3-3: P [in vitro] = 0.1668, P [in-cell] = 0.6241). (F) A7r5 cells were transfected with PPP5C siRNA for the specified incubation time and protein levels were determined using immunoblots. Statistical significance in the changes of protein levels was determined based on data from five independent replicates, and the means are presented as bar plots. The P-value was calculated using a two-tailed test with a 95% confidence interval and determined using PRISM/GraphPad version 6 (PP5: ***P = 0.001, **P = 0.0045, ***P = 0.0006; BAG3: **P = 0.0097, *P = 0.046, *P = 0.0328). (G) Smooth muscle cells were subjected to treatment with control siRNA or PP5-targeting siRNA (siPP5) followed by treatment with BafA1 to inhibit CASA mediated BAG3 degradation. Statistical significance of changes in protein levels was assessed using data from seven independent replicates, represented as boxplots. P-values were calculated using a two-way ANOVA and analyzed using PRISM/GraphPad version 6.
Source data are available for this figure.