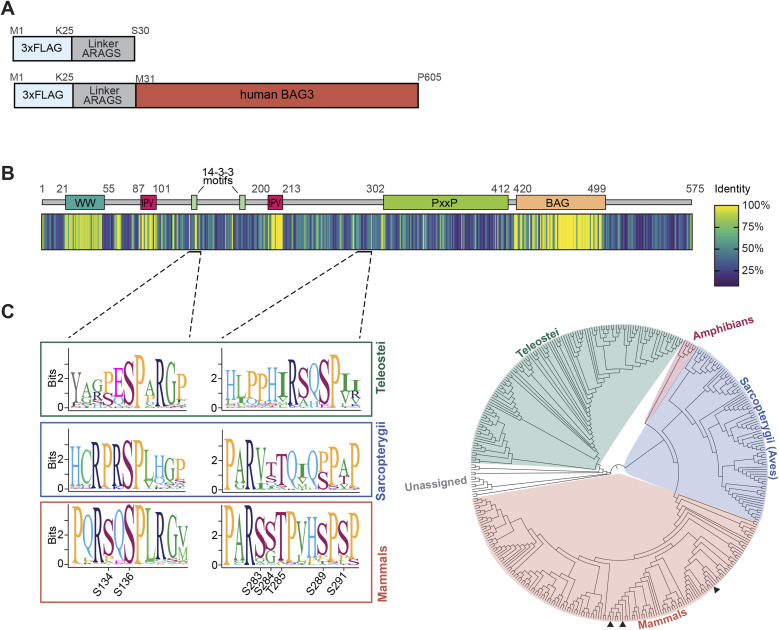
Figure S1. (Related to Fig 1). Sequence conservation of BAG3 orthologs in vertebrates.
(A) Illustration of the used FLAG-control construct to determine unspecific binding and the N-terminally triple-FLAG–tagged human BAG3 used as the bait for the FLAG co-IP. (B) A schematic overview displays annotated BAG3 domains for orientation. BAG3 sequences available from public databases (377 species, all vertebrates) were aligned by geneious using the MUSCLE algorithm. A heat map representation of the alignment identity score was generated to visualize the sequence conservation of BAG3. Regions in the alignment corresponding to gaps in the reference (BAG3 of homo sapiens) were excluded. (C) The multiple sequence alignment for the p-site stretches of interest in this study are underlined and represented in more detail using sequence logos (see (C)). (B, C) Phylogenetic analysis of the multiple sequence alignment displayed in (B). A consensus tree was built with Jukes-Contor model and Neighbor-Joining method after 200 bootstrap replications. The consensus tree for the 377 BAG3 orthologs reveals three primary clusters, corresponding to Teleostei, Sarcopterygii, and mammals. Sequence logos display the context of BAG3-pS136 and the p-site cluster (pS284-pS291) phosphorylation sites for these three principal orthologous groups. Phosphorylation sites are not conserved across all vertebrate classes, but they are conserved in mammals. In the tree, the BAG3 positions of mouse, rat, and human are indicated by black triangles, arranged from left to right.