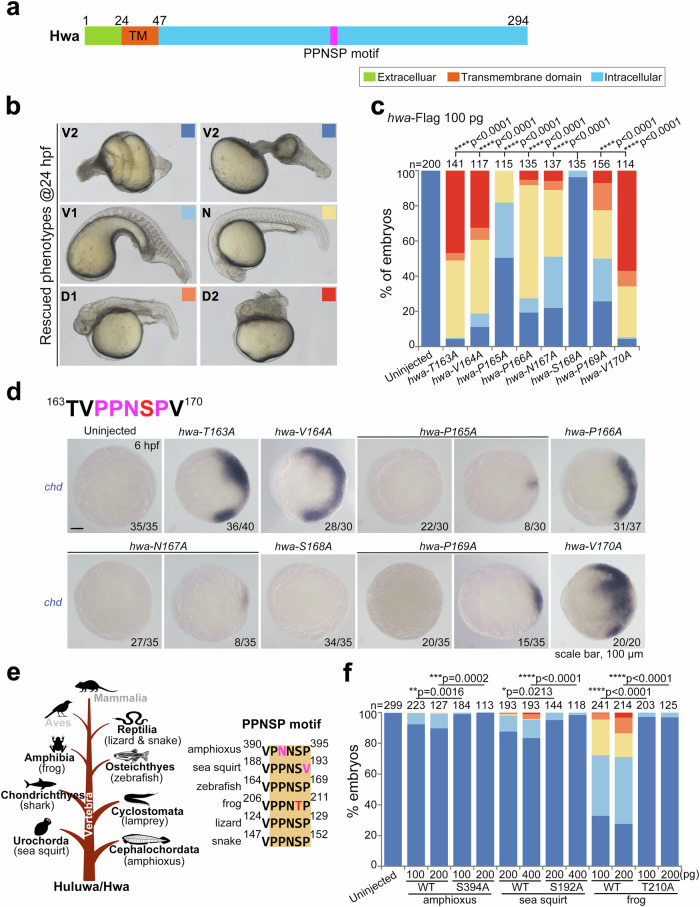
Fig. 1. Ser168 is indispensable for the axis-inducing activity of Hwa.
a Domain and motif composition of zebrafish Hwa protein. b Phenotypes of Mhwatsu01sm/tsu01sm embryos rescued by different mRNAs. These were grouped into five classes from ventralized to dorsalized at 24 hpf (V2 < V1 < N < D1 < D2; V, ventralized; N, normal; D, dorsalized), indicated by squares with different colors. c The rescue efficiency of different point-mutated mRNAs of hwa (163-170Aa) was plotted according to the classification criteria in (b), N = 2. d The upper panel shows the 163-170 aa sequence of Hwa, including the conserved PPNSP motif; the lower panel presents the expression of the dorsal marker gene chd in Mhwatsu01sm/tsu01sm embryos rescued by different mutant mRNAs of hwa, injected at the 1-cell stage and harvested at 6 hpf. All figures were imaged from the animal view, with numbers at the right corner. Scale bar, 100 μm; n, numbers of embryos. e The left panel indicates the presence of Hwa protein in the phylum Chordata excluding two classes (Aves and Mammalia are marked in gray color); the right panel shows the conservation of the PPNSP motif in different species. f The rescue efficiency of wild type (WT) and point-mutation hwa mRNAs from different species in zebrafish Mhwatsu01sm/tsu01sm embryos according to the classification criteria in (b), N = 3. 100 pg of each hwa-Flag mRNA (b–d) was injected per embryo at the 1-cell stage. (c–f) A two-tailed Fisher’s exact test was performed to evaluate differences between treatments (all phenotypes were divided into two groups: Unchanged [V2] and Changed [V1-D2]). N, number of biological replicates; n, total number of embryos in each treatment; Significant differences are indicated by ns ≥ 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Source data are provided as a Source Data file.