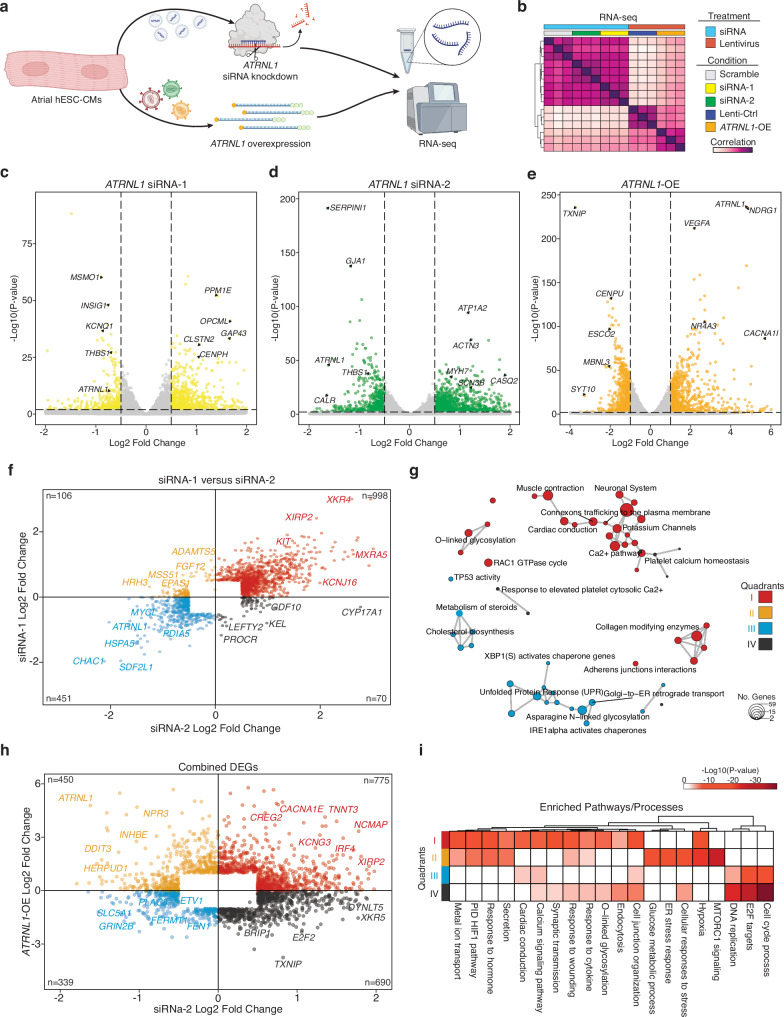
Fig. 4. ATRNL1 is part of the CM cell stress response.
a Experimental overview for RNA-seq. Created partly in BioRender. Hill, M. (2021) BioRender.com/e34r263. b Correlation heatmap of RNA-seq sample distances. Euclidean distance between samples based on DESeq2 normalized gene expression (“Methods”). Individual libraries are annotated type of treatment and condition. We profiled 3 replicates for each condition (total n = 15). c Volcano plot displaying the differentially expressed genes in hESC-aCMs between siRNA-1 treated cells and Scramble siRNA control treated cells. Genes highlighted in yellow are significantly differentially expressed (adjusted p-value < 0.05, and Log2 Fold Change > 0.5). d Volcano plot displaying the differentially expressed genes in hESC-aCMs between siRNA-2 treated cells and Scramble siRNA control treated cells. Genes highlighted in green are significantly differentially expressed (adjusted p-value < 0.05, and Log2 Fold Change > 0.5). e Volcano plot displaying the differentially expressed genes in hESC-aCMs between ATRNL1-OE treated cells and control GFP lentiviral treated cells. Genes highlighted in orange are significantly differentially expressed (adjusted p-value < 0.01, and Log2 Fold Change > 1.0). f Scatterplot of all combined significantly differentially expressed genes compared across siRNA-1 treated cells and siRNA-2 treated cells. Individual genes are colored by their location on the Cartesian coordinate plane, separating them into quadrants. Representative genes are highlighted. g Enrichment map for gene pathway over-representation analysis colored by quadrant from Fig. 5f. The size of each dot represents the number of genes in that pathway category. h Scatterplot of all combined significantly differentially expressed genes compared across siRNA-2 treated cells and ATRNL1-OE cells. Individual genes are colored by their location on the Cartesian coordinate plane, separating them into quadrants. Representative genes are highlighted. i Heatmap for pathway enrichment analysis colored by significance of enrichment (2-sided P-value). Quadrant are annotated and colored according to Fig. 5h. Volcano plots show log fold change and two-sided P-value expression changes between AF and CTRL for each gene tested using DEseq2 differential expression analysis (“Methods”).