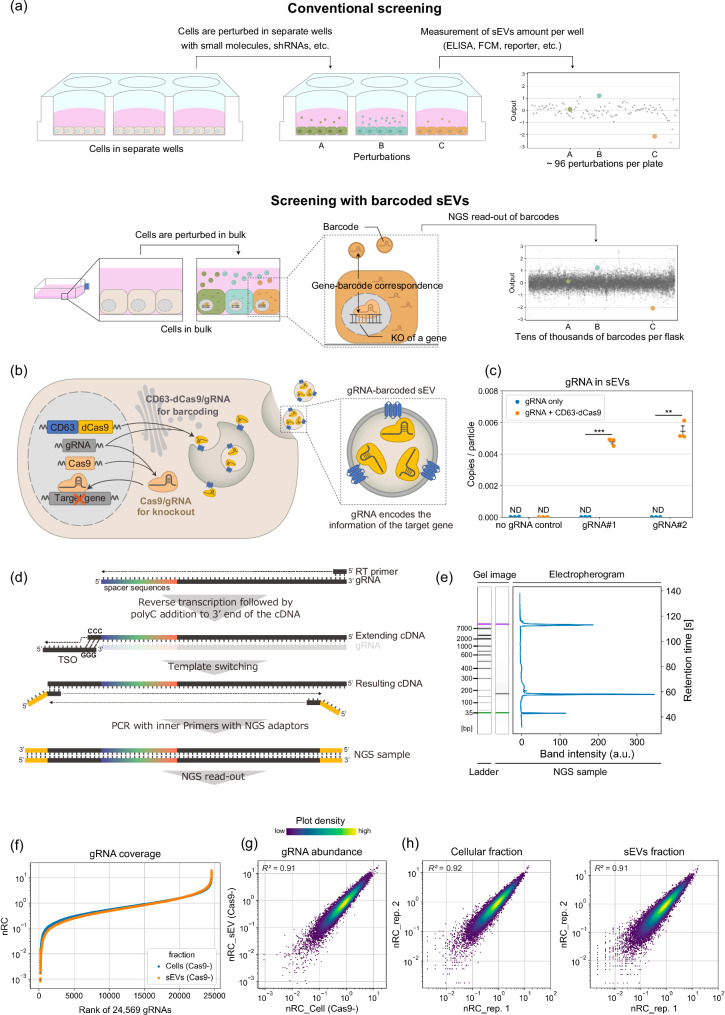
Fig. 1. Concept and creation of gRNA-barcoded sEVs for high-throughput analyses of their release regulators.
a Schematic illustration of the use of “barcoded” sEVs. Conventional biological assays (e.g., by treating cells in separate wells and measuring the amount of sEVs released in each well) offer only rather low throughput. On the other hand, sEVs barcoded with CRISPR gRNA allow for the identification of sEV release regulators in a pooled manner. b Schematic illustration of the generation of sEVs barcoded with gRNA. By expressing a fusion protein of CD63 (sEV marker) and dead Cas9 (dCas9) together with Cas9 in the sEV-producing cells, gRNA used for gene knockout can be actively encapsulated in sEVs. c gRNA abundance in sEVs isolated from culture media of HEK293T or HEK293T cells expressing CD63-dCas9 stably transduced with gRNA using lentivirus. The copy numbers of gRNAs were determined by qPCR and divided by the particle number. The Ct values of qPCR for gRNA#1 and #2 without CD63-dCas9 were not significantly different from the no-gRNA control (shown as ND: not determined). p: one sample t-test under the null hypothesis where copies/particle equals 0. **p < 0.005, ***p < 0.0005. Error bars represent ± SEM of biological replicates (n = 3). d Schematics of the developed method for spacer amplification for next-generation sequencing (NGS). e A Bioanalyzer electropherogram of an NGS sample prepared from sEV RNA as shown in Fig. 1d. f Coverage of gRNAs. Cells expressing CD63-dCas9 were transduced with a library of 24,569 gRNAs using lentivirus (DTKP library). RNAs extracted from cells and sEVs released from them were processed for NGS read-out. Read counts for every single gRNA were divided by the total sample count, then multiplied by the number of gRNAs in the library (24,569) to calculate the normalized read count (nRC), such that the mean value becomes 1. Data are shown as mean nRC from two biological replicates. g Correlation between nRCs of 24,569 gRNAs in sEVs and cellular fractions from 2 replicate cultures of Cas9− cells. h Reproducibility of nRCs of 24,569 gRNAs from cellular and sEVs fractions measured by NGS.