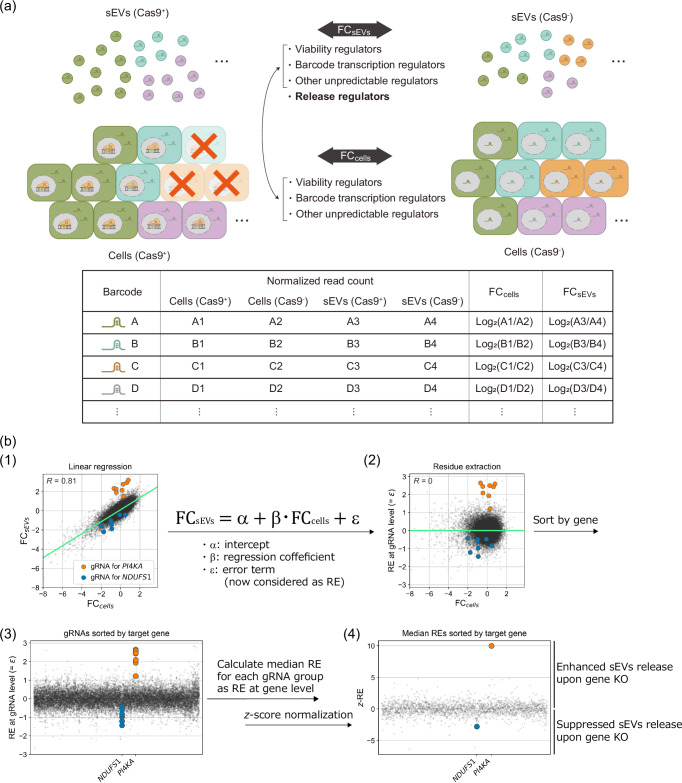
Fig. 2. Procedure of CIBER screening.
a HEK293T CD63-dCas9 cells with/without Cas9 are lentivirally transduced with the gRNA libraries, and the RNA is harvested from both cellular and sEV fractions of these cells. After NGS analysis of the gRNA composition in each sample, FCsEVs and FCcells (FC stands for fold change) of each gRNA are calculated as described. The FCcells of each gRNA reflects the change in the abundance of each gRNA in cells due to the KO of viability regulators, barcode transcription regulators, etc. FCsEVs reflects the effect of sEV release regulators in addition to the factors affecting FCcells. Therefore, the actual contribution of each gRNA (and gene) to the change of sEV release can be estimated from both FCsEVs and FCcells by means of the following procedure. b Calculation of z-normalized release effect (z-RE) from raw gRNA read counts. (1) Each gRNA was plotted with FCsEVs on the y-axis and FCcells on the x-axis. A linear relationship between FCcells and FCsEVs is observed. The regression line is shown in green (the results for the DTKP library, consisting of 24,569 gRNAs (see Supplementary Fig. 7), are shown: R = 0.81). For clarity, gRNAs targeting PI4KA and NDUFS1, which were subsequently validated as true hits, are highlighted in different colors. (2) Residue values of each gRNA obtained by performing linear regression on (1) are displayed as RE at the gRNA level. (3) gRNAs are sorted according to their target genes. (4) z-RE of each gene is calculated by taking the median value of RE at the gRNA level.