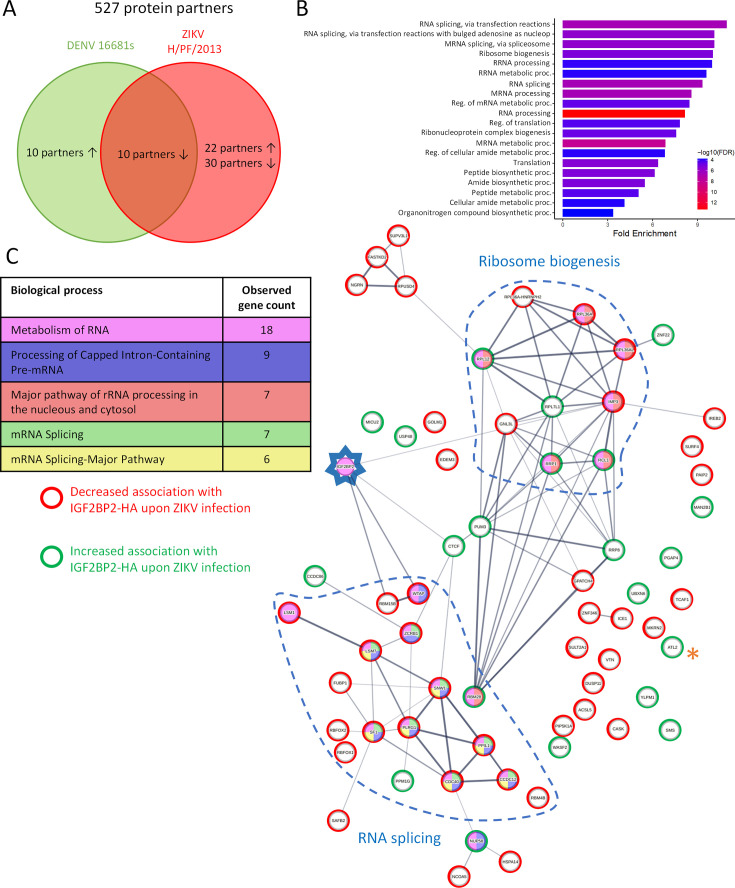
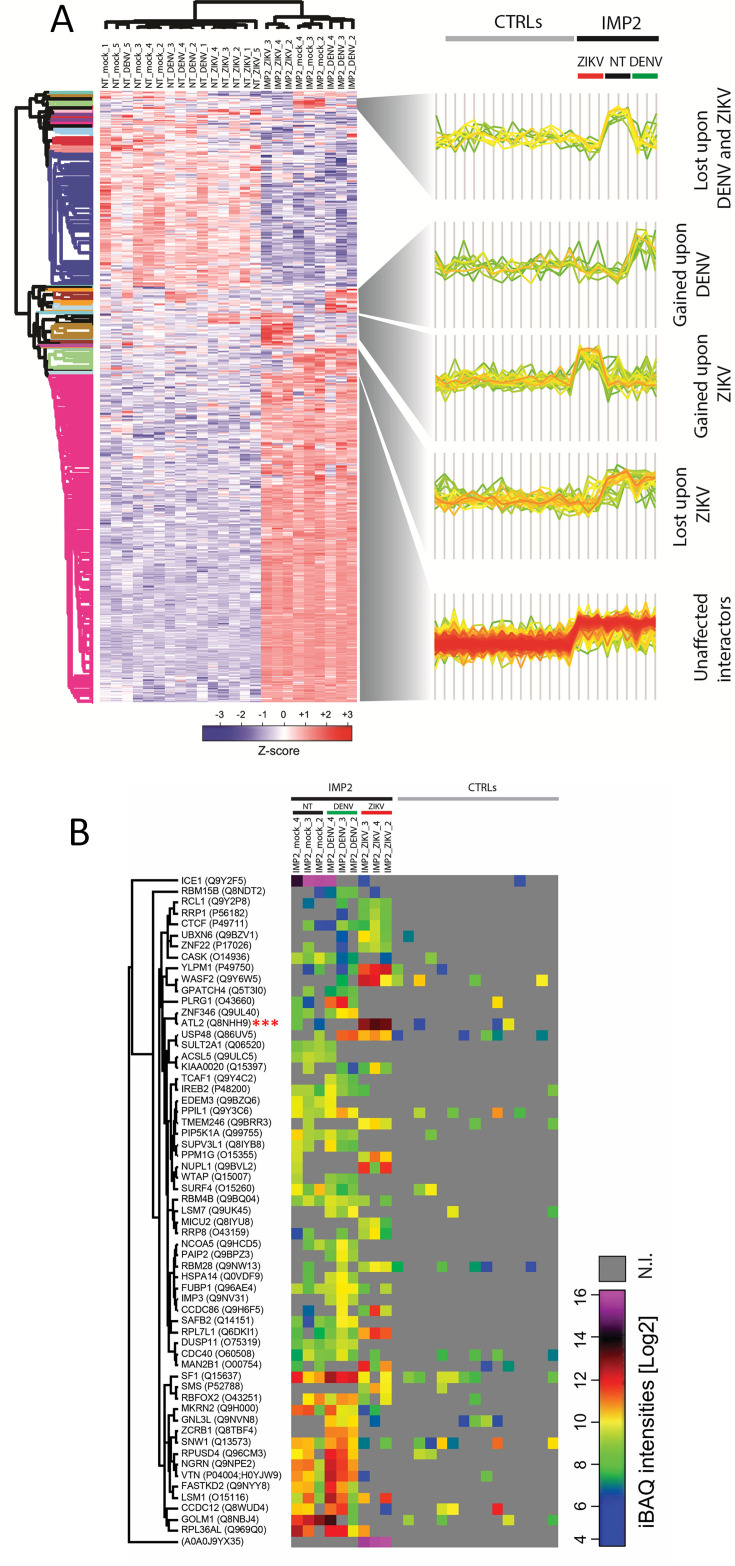
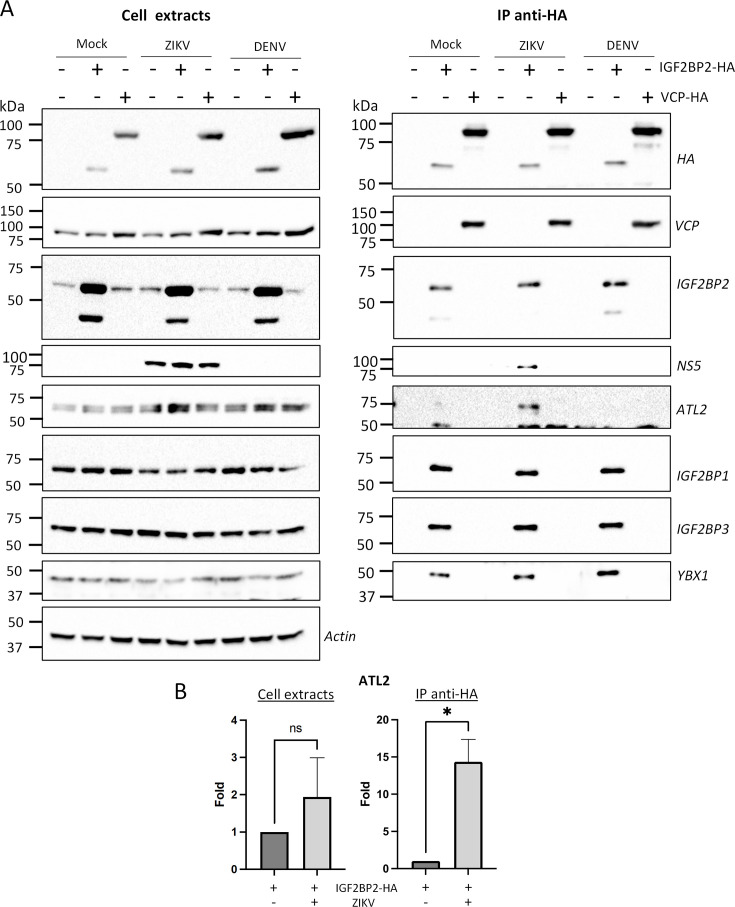
Figure 9. Zika virus (ZIKV) infection alters the IGF2BP2 proteo-interactome.
Huh7.5 cells expressing IGF2BP2-HA and control cells were infected with ZIKV H/PF/2013, dengue virus serotype 2 (DENV2) 16681s, or left uninfected. Two days later, cell extracts were prepared and subjected to anti-HA immunoprecipitations. Resulting complexes were analyzed by quantitative mass spectrometry. (A) Venn diagram depicting the overlap between IGF2BP2 partners modulated by ZIKV and/or DENV infections. (B) Gene ontology (GO) biological process analyses of the IGF2BP2 interactions which were impacted upon ZIKV infection. (C) Interaction tree of the 62 IGF2BP2 interactions modulated by ZIKV infection (generated with STRING online resource). The red and green circles identify the partners of the STRING network whose association with IGF2BP2 is decreased and increased during infection, respectively. The biological process analysis generated by STRING is also shown.