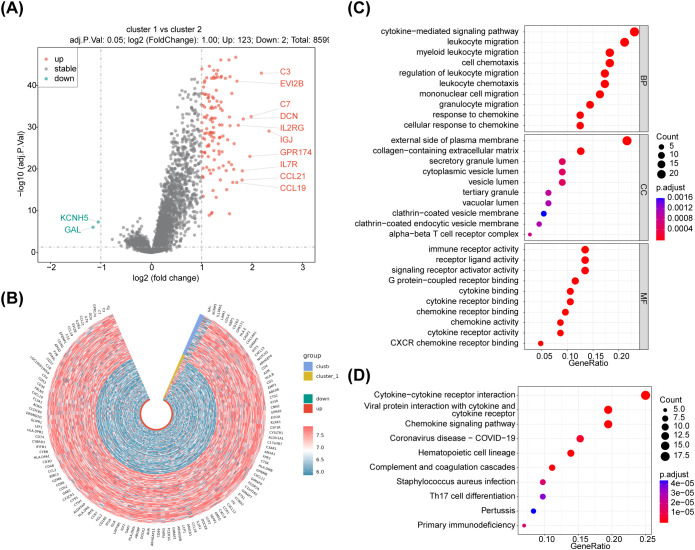
Figure 2.
Identification of differentially expressed genes associated with NETs, were plotted using R package ggpubr and pheatmap, enrichment analysis of DEG, respectively. (A) The volcano maps, there were 125 differentially expressed genes between cluster 1 vs cluster 2 groups, of which 123 were up-regulated expressed genes and 2 were down-regulated expressed genes. (B) The heatmaps,the red dots on the graph indicate up-regulated genes, the blue dots indicate down-regulated genes, the black ones are non-significant, and the more skewed towards the upper left and right corners of the graph, the greater the multiplicity of differences in significance. (C) GO and (D) KEGG enrichment analyses,125 NETs-related differential genes for GO functional annotation.