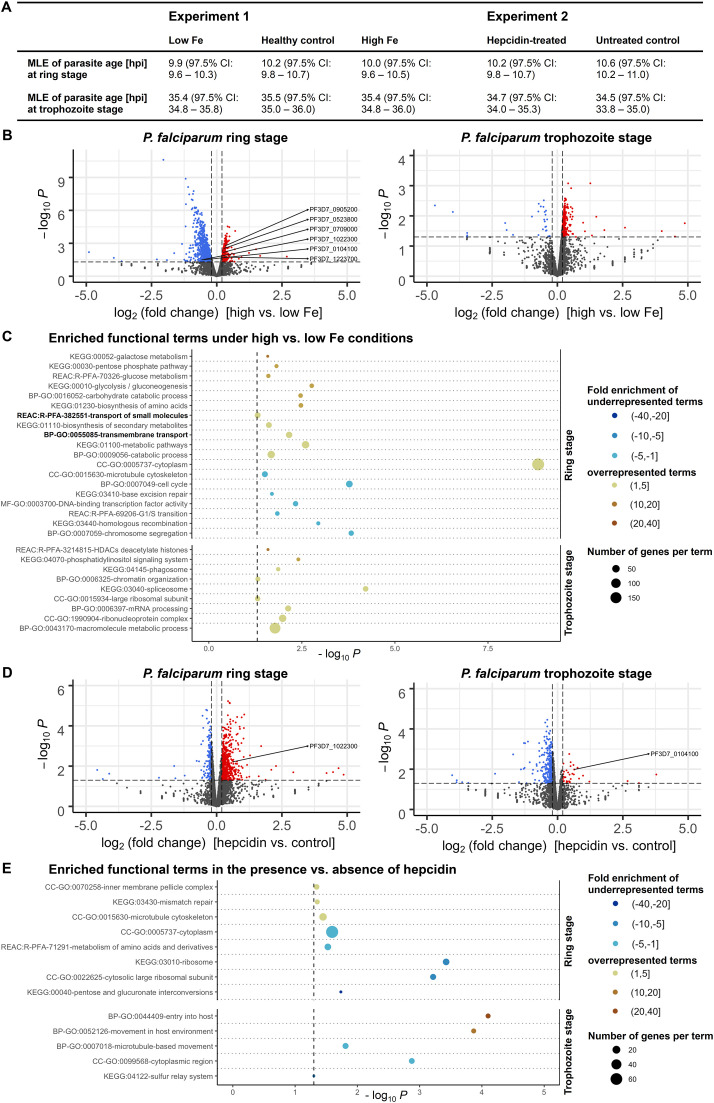
Figure 2.
Differential expression of P. falciparum 3D7 genes under various iron conditions. Parasites were cultured with erythrocytes from an individual with high, medium (healthy) or low iron status (experiment 1) or with red blood cells from another healthy donor in the presence or absence of 0.7 µM hepcidin (experiment 2). Samples were harvested at the ring and trophozoite stage (6 – 9 and 26 – 29 hours post invasion, hpi) with three biological replicates per time point and condition. The maximum likelihood estimation (MLE) of the average developmental age of the parasites for each condition and time point (A) was calculated using an algorithm developed by Avi Feller and Jacob Lemieux (Lemieux et al., 2009). CI, confidence interval. The volcano plots (B, D) show transcriptional changes of all parasite genes. Red dots indicate significantly (P < 0.05, exact test for negative binomial distribution) upregulated genes (log2 (fold change) ≥ 0.2), blue dots stand for significantly downregulated genes (log2 (fold change) ≤ -0.2), while grey dots represent genes that did not significantly differ in transcription under the conditions described (P ≥ 0.05 and/or -0.2 < log2 (fold change) < 0.2). Differentially expressed genes encoding putative iron transporters (see Table 1 ) are labeled. Panels (C, E) show the enrichment of Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome (REAC) terms among significantly regulated genes excluding var, stevor and rifin gene families at the two time points. The functional terms were summarized using REVIGO (Supek et al., 2011) to remove redundancies, represented by circles and plotted according to the significance of their enrichment (-log10 (adjusted P), hypergeometric test). The size of the circle is proportional to the number of differentially regulated genes in the dataset that are associated with the respective term, while the color stands for the fold enrichment. The gray dashed line indicates the threshold of the adjusted P value (-log10 0.05 = 1.3).