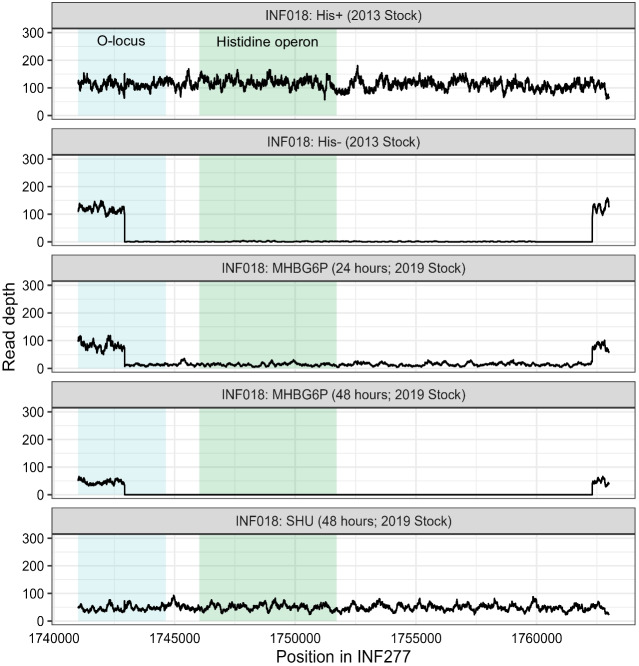
Fig. 2.
Read mapping of experimentally verified INF018 His+ and His− strains (2013 frozen stock) as well as INF018 reads from the in vitro bladder model experiment (accessions: SAMN17846711, SAMN17846713 and SAMN21465650) [12] wherein INF018 (2019 frozen stock) was passaged in cation-adjusted Mueller–Hinton broth (MHBG6P) or synthetic human urine (SHU), prior to sequencing at 24 and 48-h time-points. High-level Fosfomycin-resistant (HLR) subpopulations from each passage were also sequenced [12]. Reads were aligned to K. pneumoniae INF277 (accession: SAMN06112222, His +), which is part of the same transmission cluster as INF018 [5]. The green box highlights the location of the L-histidine biosynthesis operon, and the blue box highlights the 3′ region of the O-antigen locus. Mean sequencing depth for 2013 INF018 genomes: His+ (108.9x), His.− (113.88x); 2019 INF018 genomes: MHBG6P 24 h (82.11x), MHBG6P 48 h (49.48x), SHU 48 h (52.27x)