Abstract
African swine fever virus (ASFV), which poses significant risks to the global economy, encodes a unique host-independent transcription system. This system comprises an eight-subunit RNA polymerase (vRNAP), temporally expressed transcription factors and transcript associated proteins, facilitating cross-species transmission via intermediate host. The protein composition of the virion and the presence of transcription factors in virus genome suggest existence of distinct transcription systems during viral infection. However, the precise mechanisms of transcription regulation remain elusive. Through analyses of dynamic transcriptome, vRNAP-associated components and cell-based assay, the critical role of M1249L in viral transcription regulation has been highlighted. Atomic-resolution structures of vRNAP-M1249L supercomplex, exhibiting a variety of conformations, have uncovered the dual functions of M1249L. During early transcription, M1249L could serve as multiple temporary transcription factors with C-terminal domain acting as a switcher for activation/inactivation, while during late transcription it aids in the packaging of the transcription machinery. The structural and functional characteristics of M1249L underscore its vital roles in ASFV transcription, packaging, and capsid assembly, presenting novel opportunities for therapeutic intervention.
Subject terms: Cryoelectron microscopy, Virus structures
African swine fever virus (ASFV) is a contagious DNA virus causing lethal disease in swine. This study reveals structures of ASFV RNA polymerase bound to protein M1249L, showing its vital roles in regulating early transcription and packaging.
Introduction
African Swine Fever virus (ASFV), a member of the Asfarviridae family, is the only known large DNA arbovirus and classified as nucleocytoplasmic large DNA virus (NCLDV), which also includes the canonical vaccinia virus (VACV)1. ASFV is the causative agent of African Swine Fever (ASF), a highly contagious disease with a mortality rate of up to 100%. The absence of effective vaccines or treatments makes ASF a significant threat to the global economy. The structure of ASFV is complex, comprising five layers: an outer envelope, capsid, inner envelope, inner core shell, and nucleoid2. Cryo-electron microscopy studies have revealed that the viral capsid is primarily composed of the major capsid protein p72 and four minor capsid proteins M1249L, p17, p49, and H240R3. ASFV primarily invades host cells through dynamin-dependent and clathrin-mediated endocytosis, as well as macropinocytosis4,5. Following entry into the cytoplasm, early ASFV transcription is initiated using enzymes and factors that are pre-packaged within the virions6. While there is a brief nuclear phase, the majority of the replication process and all viral morphogenesis occur within the cytoplasm of the infected cell7,8.
VACV and ASFV share several notable characteristics, including double-stranded DNA genomes of 170-200 kilobases, cytoplasmic replication, and various structural similarities. ASFV dedicates approximately 20% of its genome to encode the genes involved in viral mRNA transcription and modification, exhibiting precise spatial and temporal regulation of ASFV gene expression independent from its host transcription system9,10. Like most NCLDV, ASFV transcription can be divided into four stages: immediate-early, early, intermediate, and late11. A unique feature of ASFV is its complete set of viral transcription factors, which bind to specific DNA sequences typically located in the promoter regions to regulate and initiate the viral transcription during different infection stages, thereby creating a favorable environment for its replication. This lack of dependence on host factors may also allow ASFV to spread among different breeds of pigs and its evolutionary distant tick vectors. Upon viral entry and uncoating in the cytoplasm, enzymes required for DNA replication and RNA transcription, as along with viral encoded transcription factors, are expressed. Early transcription utilizes virus-encoded transcription and transcript processing enzymes packaged within the core12. Similar to other NCLDV, ASFV uses a temporal gene expression strategy throughout the respective stages of the ASFV replication cycle10,13.
Many NCLDVs encode viral RNA polymerase (vRNAP) complexes that functions similarly to those of eukaryotes, which enables cytoplasmic transcription. To date, among NCLDVs, the architecture and functional dynamics of VACV vRNAPs have been extensively studied14–16. Notably, the VACV core vRNAP comprises eight subunits, including a TFIIS homolog, but lacks vRPB9 and TFIIB. Instead, VACV core vRNAP contains a poxvirus-specific transcription factor, Rap94, which is homologous to TFIIB and binds to the enzyme periphery16. Furthermore, bioinformatic analyses demonstrate that ASFV exhibits similar redundancy in transcription factors, indicating that multiple transcription factor systems in ASFV may play roles at different stages of the viral infection cycle, transcribing different clusters of viral proteins17. Recently, the structure of a recombinant 8-subunit RNAP core complex from insect cell was reported18, including pNP1450L (vRPB1), pEP1242L (vRPB2), pH359L (vRPB3-11), pD205R (vRPB5), pC147L (vRPB6), pD339L (vRPB7), pC122R (vRPB9) and pCP80R (vRPB10). However, due to the high pathogenicity of ASFV, experimental data regarding ASFV gene transcription and transcript processing enzymes packaging during viral assembly remain limited. Consequently, little is known about the biogenesis, assembly, mechanism, and regulation of ASFV transcription machinery. Intriguingly, questions persist about how the highly dynamic vRNAP-associated complexes required for transcription during early stages of infection are packaged into ASFV particles. Furthermore, the roles of transcription factors remain unknown, as no structure with bound transcription factors has been reported to date.
In this study, we present high-resolution cryoEM structures of ASFV vRNAP core (PDB-8YQV and EMD-39507), core complexed with endogenous DNA (PDB-8YQZ and EMD-39511), and the core complexed with M1249L (including 5 complex structures and 26 maps illustrating its conformational flexibility) that was previously recognized as a capsid component. Structural comparisons with homologous proteins and functional analyses suggest that M1249L is a multifunctional protein that not only participates in capsid assembly, also plays roles in vRNAP constitution and transcription regulation. Our findings provide new insights into the pathogenicity of ASFV and provide a framework for developing countermeasures to prevent and control ASF outbreaks.
Results
M1249L plays crucial roles for ASFV vRNAP
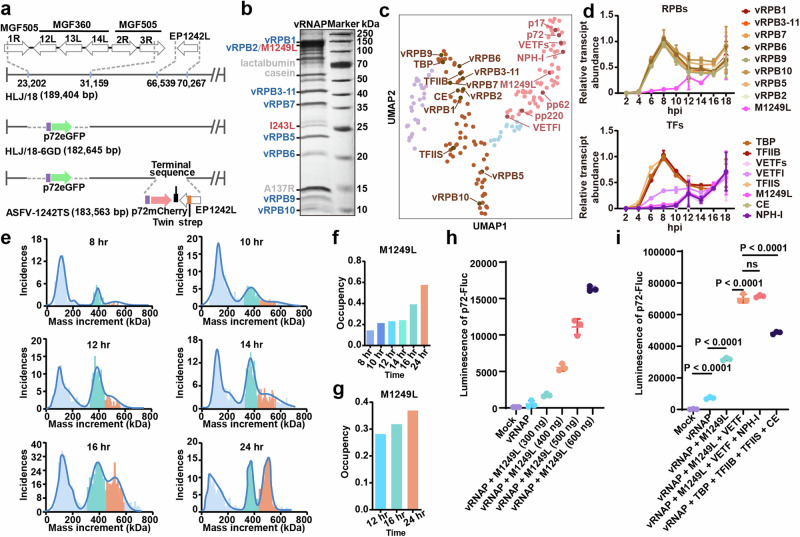
To affinity-purify the endogenous ASFV-vRNAP complex, we constructed a recombinant ASFV-1242TS virus with a C-terminal twin-strep tag on the EP1242L gene, based on the attenuated ASF vaccine candidate strain HLJ/18-6GD19 (Fig. 1a). The recombinant virus was validated by intrinsic eGFP/mCherry fluorescence and Western Blot analysis (Supplementary Fig. 1a, b), and shows a similar growth curve to native HLJ/18-6GD in porcine alveolar macrophages (PAM) (Supplementary Fig. 1a). This confirmed that the addition of the affinity tag to EP1242L does not affect viral replication capacity. For the expression of vRNAP, porcine bone marrow (PBM) cells were infected. The vRNAP complex was then purified using Strep-Tactin XT resin with a yield of ~0.1 mg from 1 liter of PBM cells at 48 hrs post-infection (hpi) at a multiplicity of infection (MOI) of 1. SDS-PAGE analysis, followed by silver-staining and MALDI-TOF mass spectrometry, confirmed the presence of the eight vRNAP subunits (Fig. 1b). To further investigate the dynamics of ASFV vRNAP, we purified vRNAP complexes from PBM cells at various time points (8/10/12/14/16/24 hpi). Silver staining indicated that M1249L was consistently co-purified with the vRNAP subunits, with an increasing proportion in the purified samples obtained at 12/16/24 hpi (Fig. 1b).
Fig. 1. M1249L plays a crucial role in the activity of ASFV vRNAP.
a Schematic representation of modified ASFV genes. A DNA fragment encoding a Twin strep tag was fused in HLJ/18-6GD to the 3’ end of EP1242L, allowing the expression of C-terminally tagged vRPB2. b Proteins eluted from XT beads visualized by silver staining on a 12.5% SDS-PAGE. RNA polymerase eight subunits are colored blue, transcription factors are colored red, and pollutant proteins are colored gray. Protein bands were characterized by the molecular weight marker and identified by MS Fingerprinting analysis, with protein names indicated on the left next to their corresponding bands. The experiment was repeated independently twice with similar results. c UMAP analysis of the ASFV transcriptome demonstrates expression patterns of vRNAP and transcription factors across various stages. Purple dots denote immediate early-expressed genes, brown dots denote early-expressed genes, blue dots denote mid-term expressed genes and pink dots denote late-expressed genes. d The transcriptome analysis of ASFV vRNAP subunits plus M1249L (top) and transcription factors (bottom) at different time points post-infection (n = 3). Data are presented as mean values +/- SD. e Measurement of vRNAP complex molecular weight at different time points by mass spectrometry. The peak of core RNAP is colored cyan, while the vRNAP-M1249L complex is colored orange. f The proportion of vRNAP-M1249L complexes determined by mass spectrometry (top) as well as silver staining (bottom) relative to the total particles in the sample g. h HEK293T cells were transfected with plasmids of p72-Fluc and varying doses of M1249L, the luminescence was detected by luminometer (n = 3). Data are presented as mean values +/- SD. i HEK293T cells were transfected with p72-Fluc with or without certain transcription factor, the luminescence was detected by luminometer (n = 3). Data are presented as mean values +/- SD. Statistical analysis was performed by unpaired t test. P values are marked. Source data are provided as a Source Data file.
UMAP analysis of the dynamic ASFV transcriptome shows that the virus-encoded transcription factors NP868R (CE), I243L (TFIIS), C315R (TFIIB), and B263R (TBP) cluster with vRNAP subunits as early transcribed genes, peaking at 8 hpi, while transcription factors D1133L (VETFs), G1340L (VETFl), Q706L (NPH-I), virion structural proteins p17/p72/pp62/pp220 and M1249L are grouped in a separate cluster as late transcribed genes peaking at 12 hpi (Fig. 1c, d). Considering the presence of VETF, NPH-I, and M1249L in purified ASFV particles along with vRNAP subunits20, and given the involvement of VETF and NPH-I in VACV vRNAP complex formation14,16, we deduce that M1249L may also contribute to ASFV transcription. Mass photometry measurements indicated that purified ASFV vRNAP primarily exists in two peaks, with molecular weights of approximately 400 kDa and 540 kDa. Given the molecular weight of M1249L ( ~ 144 kDa), these peaks correspond to the ASFV eight-subunit core RNAP and RNAP-M1249L complex, respectively (Fig. 1e). Furthermore, mass photometry measurements demonstrate that the proportion of vRNAP-M1249L complex gradually increased in 8/10/12/14/16/24/48 hr samples (Fig. 1f), consistent with the results of silver staining analysis (Fig. 1g and Supplementary Fig. 1c).
To further investigate the function of M1249L, we employed a previously reported luciferase reporter system, in which the luciferase downstream of p72 promoter is transcribed by the co-transfected ASFV RNAP21, along with other transcription factors. We found that co-transfection of M1249L with vRNAP resulted in significant, dose-dependent increase in vRNAP activity (Fig. 1h), indicating that M1249L plays vital roles in ASFV transcription. Additionally, various combinations of transcription factors were co-transfected with vRNAP and M1249L, and the results showed that the inclusion of VETF enhanced the transcriptional activity of the vRNAP-M1249L complex, while NPH-I had no additional effect (Fig. 1i). In contrast, the addition of TBP-TFIIB-TFIIS-CE transcription factors to vRNAP also enhanced transcriptional activity, though not as significantly as the M1249L + VETF or M1249L + VETF + NPD-I combinations (Fig. 1i).
Structure determination of ASFV core vRNAP and vRNAP-M1249L complex
To investigate the impact of M1249L on the regulatory activity of vRNAP, we purified the ASFV-vRNAP complex at 48 hpi and obtained cryo-EM reconstructions of two states of the vRNAP complex at resolutions of 2.6 Å (PDB: 8YQT) and 2.7 Å (PDB: 8YQV). To elucidate the structural details of M1249L, three rounds of 3D classification were employed, producing three distinct reconstructions at resolutions of 2.6 Å (PDB: 8YQW), 2.8 Å (PDB: 8YQU), and 3.7 Å (PDB: 8YQY). Additionally, a local refinement targeting a blurred region in the map yielded a further refined map at a resolution of 3.0 Å (PDB: 8YQX) (Supplementary Figs. 2, 3 and Supplementary Table 1). To understand the interactions between vRNAP and viral transcription factors, we analyzed ASFV vRNAP complexes isolated at various time points (12/16/24 hpi). A blurred density observed in the cleft prompted further analysis. By applying a thorough statistical 3D classification method to all collected data, we identified 25 distinct reconstructions, highlighting the various states of M1249L binding to vRNAP (Supplementary Fig. 4 and Supplementary Table 1).
Overall architecture of ASFV vRNAP complex and its dynamics
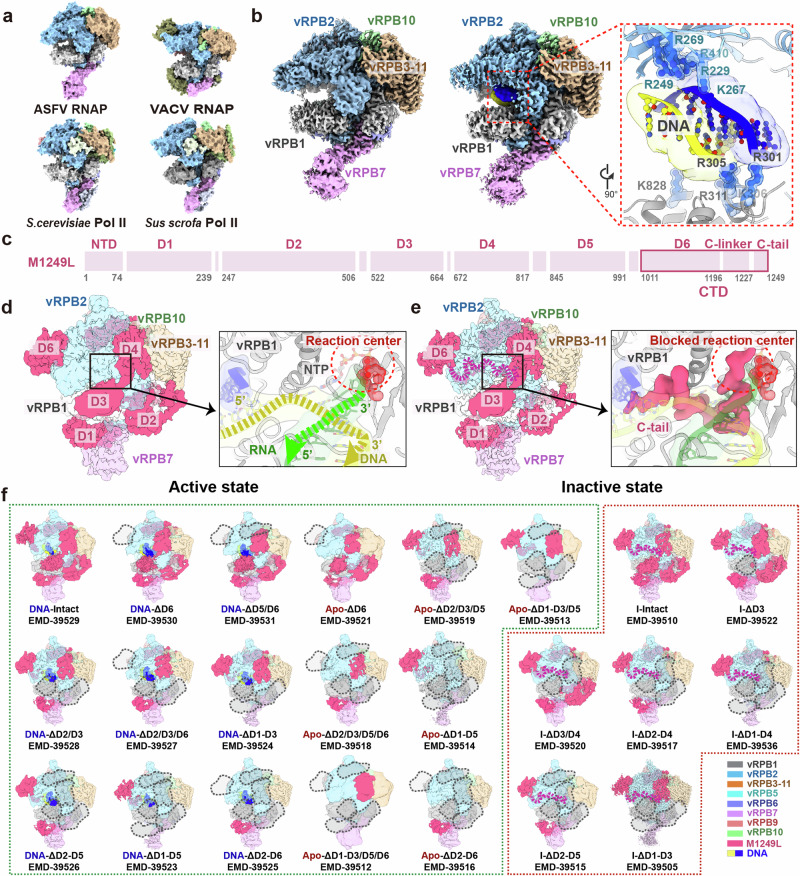
The ASFV core vRNAP (PDB: 8YQV), constituted by eight subunits, is stabilized by a large buried surface area (Supplementary Fig. 5). A comparison with previously reported insect cell-expressed recombinant vRNAP (PDB: 8Q3B) showed high similarity, with an r.m.s.d. of 0.85 Å, indicating that the structure of ASFV vRNAP is stable across different expression systems (Supplementary Fig. 5). To illustrate the unique features of ASFV core vRNAP, we superimposed vRNAPs from ASFV, VACV (PDB: 6RIC)16, yeast (PDB: 1WCM)22 and swine (PDB: 7EG7)23. Overall, ASFV vRNAP adopts a structure more closely resembling eukaryotic RNAPs than its NCLDV homolog of VACV, as confirmed by phylogenetic analysis (Fig. 2a, Supplementary Fig. 6). Additionally, we have identified endogenous nucleic acids adjacent to the active center in ~30% of vRNAP particles from combined 12/16/24 hpi purified samples. In the cleft between vRPB1 and vRPB2, next to the bridge helix at a distance of ~10 Å, an extra density of endogenous nucleic acid was observed (Fig. 2b). Consequently, we modeled an 8 bp segment of B-type double-stranded DNA (derived from PDB:7EG723) into this region. The resulting DNA model showed a good fit to the density map and formed favorable electrostatic interactions with surrounding positively charged amino acid residues, specifically R301, R305, K306, R311, K828 from vRPB1, and R229, R249, K267, R269, R410 from vRPB2. We did not detect the density of downstream/upstream DNA duplex, likely due to the high mobility of duplex DNA entering the cleft for strand separation24.
Fig. 2. Architectures of ASFV core vRNAP and vRNAP-M1249L complex.
a Comparison of ASFV vRNAP (8YQV) to S.cerevisiae (PDB: 1WCM), VACV (PDB: 6RIC), and Sus scrofa (PDB: 7EG7) depicted in surface representation. Homologous subunits are colored accordingly. b Structural comparison of core vRNAP (8YQV) with vRNAP-nucleic acid complex (8YQZ), in surface representation. We fitted a segment of B-type DNA (the yellow/blue chains represent double stranded DNA, from PDB: 7EG7) into the nucleic acids map. The closed-up view presents key amino acids involved in interaction with nucleic acid, depicted in cartoon. The labeled amino acids are color-coded to match the subunits’ colors. c Domains of M1249L, with the magenta box highlighting D6, C-linker and C-tail within C-terminal domain (CTD). d The map of vRNAP-M1249L complex without C-linker/C-tail (left), and close-up view of the active state of vRNAP (right). Colors of vRNAP subunits are consistent with (f). By adjusting transparency, density of M1249L obscured by polymerase subunits is revealed. The active site is depicted in cartoon to highlight details. The red circle indicates the position of reaction center. In active state, the active site lacks density of M1249L C-tail. Yellow and green arrows indicate the moving directions of DNA template and nascent mRNA. A nucleotide triphosphate (NTP) molecule was modeled within the NTP entry channel, depicted in red. e The map of vRNAP-M1249L complex with C-linker/C-tail (left), and close-up view of inactive state of vRNAP (right). The magenta dashed lines emphasize positions of C-linker and C-tail. In this state, density of the M1249L C-tail domain is prominently displayed, with C-terminal penetrating into the reaction center, thereby hindering binding of nucleic acids and NTP. f Dynamic variations of M1249L in two states are depicted across 23 distinct cryo-EM maps. The color scheme for each subunit is indicated at the right bottom of the panel. Green dashed box illustrates the dynamics of M1249L domains in active state. Gray dashed boxes denote the absence of domain density at that specific position of M1249L. Red dashed box signifies the dynamics of M1249L in inactive state, with magenta dashed lines emphasizing positions of C-linker and C-tail.
During 3D classification, ~50% of vRNAP was found to bind with M1249L, and its single particle reconstitution achieved an overall resolution of 3.0 Å. The structure of M1249L was modeled based on the map, except for the N-terminal 73 amino acids and the linker regions, due to their structural flexibility/dynamics. The final built M1249L structure consists of six domains (D1-D6) connected by unstructured linkers (Supplementary Fig. 3), and wraps around vRNAP at multiple sites spanning the vRNAP cleft, forming a large buried surface area of ~40,000 Å2 (Fig. 2c–e). The structure of M1249L is mainly composed of α-helices (see also Supplementary Fig. 7 and its legend for detailed structural description). Structural similarity search using DALI25 or VAST26,27 did not identify homologous structures for any domain, suggesting that M1249L bears special structural features. The vRPB1 loop, which coordinates a Mg²+ ion via NADFDGD (aa 455-461) motif containing three Asp residues, forms the catalytic center. Notably, in a subset of vRNAP particles, this catalytic center is unoccupied, allowing the accommodation of the template DNA duplex and nascent mRNA, indicating an active state (Fig. 2d). In contrast, within the remaining subset of vRNAP particles, the terminal C-tail loop (residues 1227-1249) of M1249L occupies the DNA binding site within the catalytic center and interacts with the Mg2+ ion. This interaction blocks the NTP channel, indicating that these vRNAP particles are in an inactive state (Fig. 2e).
Structural comparison indicates M1249L multi-domains function as multiple transcription factors
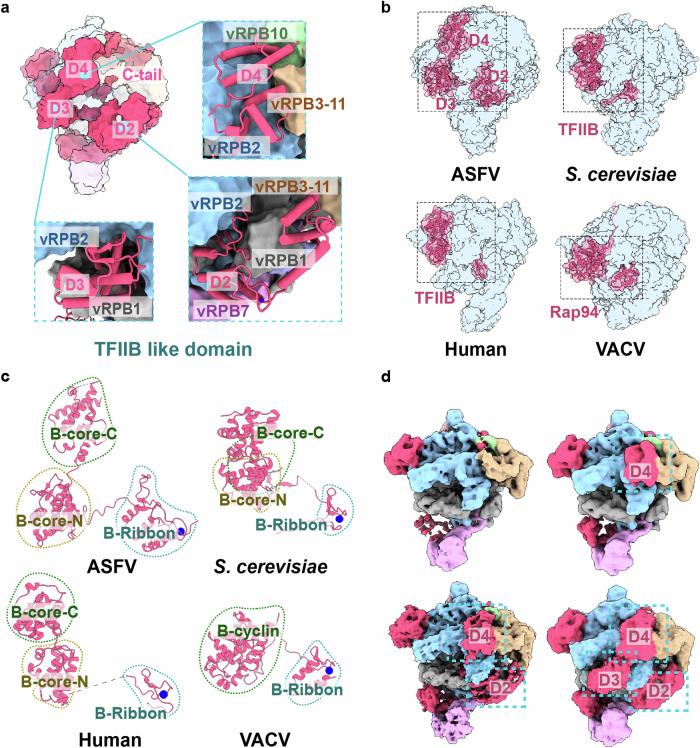
Although M1249L shares no sequence homology with VACV, the overall interaction path of M1249L on the core vRNAP, as well as the distribution of zinc-finger motifs, is similar to that of VACV Rap94 (a TFIIB-like protein), suggesting M1249L and Rap94 may have similar functions (Supplementary Fig. 7). To elucidate the roles of M1249L in transcription, we compared and modeled ASFV vRNAP-M1249L complex with structures of swine RNAP and VACV vRNAP. Based on the analysis of domain structures, binding sites, and specific amino acid motifs, we deduce that the multi-domain structure of M1249L represents an assembly of transcription factors analogs, including TFIID, TFIIB, TFIIS, and TFIIF (Figs. 3–5).
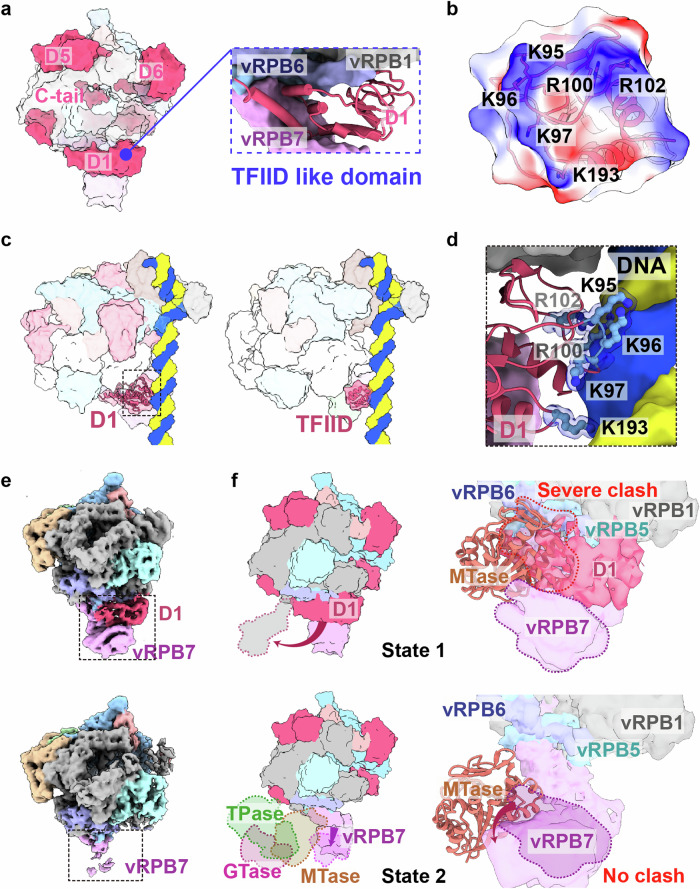
Fig. 3. Structural analysis of the M1249L D1 domain reveals its role as a transient transcription factor similar to TFIID.
a The surface structure of vRNAP-M1249L, highlighting the functional domains of M1249L, shows similarities to the classical transcription factor TFIID (see blue dashed box). b Surface potential map of M1249L D1 domain in contact with nucleic acids. c Left: ASFV RNAP-M1249L complex superposed onto Sus scrofa Pol II (PDB: 7EG7). Right: Sus scrofa Pol II (PDB: 7EG7). M1249L D1 is presented in cartoon format, with black dashed lines indicating the positions where it contacts the nucleic acid. d Details of key amino acids involved in M1249L D1-nucleic acid interactions. M1249L D1 domain depicted in cartoon, with crucial amino acids shown in ball-and-stick mode. e Two different cryo-EM maps demonstrate the stabilizing effect of M1249L D1 domain on vRPB7. f The two states of M1249L D1 domain when CE is unbound (top) or bound (bottom) to vRNAP are shown in the surface. The binding state of CE is obtained with reference to VACV CCC complex (PDB: 6RIE). The three domains of CE are colored respectively as green (TPase), magenta (GTase), and orange (MTase). The zoom-in image shows a significant clash between D1 and MTase when D1 is bound to vRNAP (top), while there is no clash when D1 moves away (bottom). The structure of the ASFV MTase is predicted by AlphaFold, displayed in cartoon format, with vRNAP shown as a surface representation.
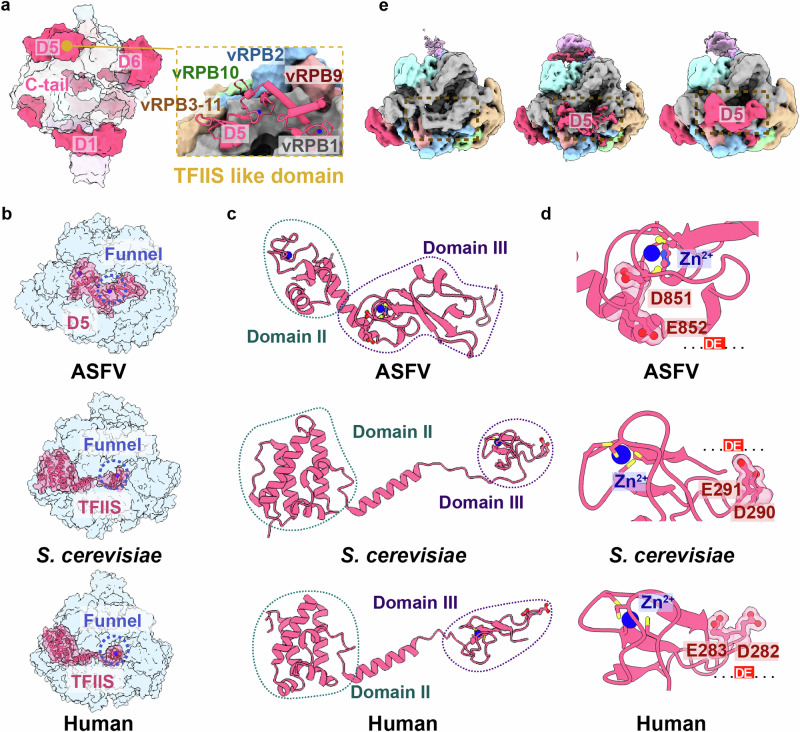
Fig. 5. Structural analysis of the M1249L D5 domain identifies it as a temporary transcription factor with similarities to TFIIS.
a The surface structure of vRNAP-M1249L, highlighting the functional domains of M1249L, shows similarities to the classical transcription factor TFIIS (yellow dashed box). b Comparison of the location of M1249L D5 domain to S.cerevisiae and human TFIIS. The whole model is shown as half-transparent blue surface with M1249L D5 domain or TFIIS shown as solid cartoon. The Zn2+ ions are shown as spheres and highlighted by a blue dashed circle. c The cartoon structures of TFIIS homolog structures from ASFV, S.cerevisiae and human. The Zn2+ ions are shown as spheres. The similar domains are indicated by dashed circles of the same color. d Close-up view shows the key acidic amino acids (…DE…) involved in functional activity with Zn2+ ions. e Three different maps illustrate the M1249L D5 domain dynamic alterations.
During transcription initiation, the interaction of RNAP II with promoter typically involves binding to essentially straight, duplex DNA28. Coincidentally, a cluster of positively charged amino acids in D1, including K95, K96, K97, R100, R102 and K193, are oriented towards the DNA backbone, poised for template binding during pre-initiation complex (PIC) assembly (Fig. 3b, Supplementary Fig. 8a). By structural superimposition with a closely related mammalian PIC (PDB: 7EG7)23, we found that D1 of M1249L is positioned upstream of the DNA template, with loops that coordinate well with the DNA grooves (Fig. 3c,d). Dynamic maps show that D1 exhibits flexibility (Fig. 3e), allowing it to be displaced during DNA bending. This flexibility is essential, as comparisons with the VACV co-transcriptional capping complex (CCC)15 indicate that the binding of D1 would sterically clash with the MTase domain within the CCC (Fig. 3f). Therefore, prior to CCC assembly, DNA bending is likely necessary to induce the translocation of D1, ensuring that the C-terminal domain (CTD) of vRPB7 remains unbound. This configuration would enable vRPB7 to subsequently associate with the capping enzyme (CE) of NP868R to form the CCC assembly at the vRPB7 C-terminal MTase small-subunit homolog (Fig. 3f, Supplementary Fig. 8b), as recently suggested by analysis of the recombinant ASFV vRNAP core18.
TFIIB plays central roles in RNA transcription initiation by recruiting Pol II to the promoter and interacting with both DNA and TBP via its N-terminal zinc-ribbon domain and C-terminal B-core cyclin fold29–32. Structural comparisons with human (PDB: 7EG7)23, yeast (PDB: 4BBR)33, and VACV (PDB: 6RFL)16 RNAPs indicate that domains D2-D3-D4 of M1249L resemble the binding location and architecture of canonical TFIIB (Fig. 4a, b). D2 contains a zinc-finger motif similar to the B-ribbon of TFIIB, followed by a B-reader/linker loop that probes to the interface between vRPB1 ‘dock’ and vRPB2 ‘hybrid binding’ domains. Meanwhile, D3 and D4 resemble the B-core cyclin fold as found in eukaryotic RNAPs (Fig. 4c). Notably, D3 is located at the RNA exit site and must be displaced to avoid clashing with the elongating RNA30,34. This is consistent with the low occupancy of D3 ( ~ 1%) and the dynamic maps showing that D2/D3/D4 are absent in various combinations (Fig. 4d). These observations suggest that during viral transcription, the TFIIB-like domains in M1249L undergo major conformational adjustments. However, in VACV, the B-core-C is replaced by an unstructured strand (Fig. 4b,c), indicating that VACV is more evolutionarily distant.
Fig. 4. The M1249L D2-D4 domains, through structural analysis, are identified as a temporary transcription factor analogous to TFIIB.
a The surface structure of vRNAP-M1249L, highlighting the functional domains of M1249L, shows similarities to the classical transcription factor TFIIB (cyan dashed box). b Comparison of the location of M1249L TFIIB-like domains to human, S.cerevisiae TFIIB and VACV Rap94 B-homology region. The whole model is shown as a transparent blue surface with TFIIB or TFIIB-like domain shown in cartoon. c The four structures of TFIIB shown in the cartoon. The Zn2+ ions in Zinc-finger motifs are shown as blue spheres. The similar domains are indicated by dashed circles of the same color. d Four maps illustrate the M1249L TFIIB-like domain dynamic alterations.
Manual structural comparison and sequence analysis reveals that D5 exhibits certain similarity to TFIIS of RNAPs from human (PDB: 8A40)35 and yeast (PDB: 1Y1V)24. Similar to eukaryotic TFIIS, D5 binds to the exterior rim of the funnel (Fig. 5a), and is constituted by a helix bundle resembling Domain II of TFIIS and a zinc-ribbon fold analogous to Domain III of TFIIS (Fig. 5b, c). Additionally, D5 also includes an acidic hairpin containing two consecutive acidic residues (D851 and E852), which are strictly conserved in TFIIS (Fig. 5d) and critical for elongation and RNA cleavage36. However, the binding site of D5 does not perfectly coincide with that of canonical eukaryotic TFIIS37, and the position/orientation of the acidic hairpin do not fully align, suggesting that D5 may be in an untriggered state, poised to move into the funnel only when elongation or backtracking is required. This hypothesis is supported by the dynamic maps, which show that the acidic hairpin density is relatively weak and flexible (Fig. 5e).
Discussion
The vRNAP complex plays crucial roles during ASFV infection and proliferation, participating in the synthesis of viral mRNA, RNA genome replication (as in RNA viruses), virus particle assembly, and viral genome package38. By adding an affinity tag to subunit vRPB2, we isolated the endogenous ASFV core vRNAP and its supercomplex with M1249L. The ASFV core vRNAP closely resembles host RNAP-II, suggesting that ASFV vRNAP is evolutionary conserved which is confirmed by phylogenetic analysis (Supplementary Fig. 6b). ASFV-encoded M1249L is a late-phase protein9 and a skeleton protein with fiber-like zipper structure participating in capsid assembly3. Interestingly, when we compare the map/structure of M1249L in the vRNAP to that in the viral capsid, no structural similarities were observed (Supplementary Fig. 10). This discrepancy may be due to M1249L forming extensive intermolecular networks with other capsid proteins(e.g. p72 and p17), which induce the conformation of M1249L as a zipper-like structure. Currently, knowledge about M1249L is limited, though previous reports suggest that M1249L may also participate in regulating host immune signaling pathways by antagonizing type I interferon39,40.
ASFV encodes virus-specific and host-homologous transcription factors involved in initiation, elongation, and termination, e.g. B263R as TBP, C315R as a TFIIB-like factor, and I243L as an elongation factor TFIIS17. This indicates that ASFV has a host-independent transcription system, enabling it to survive in different hosts. We deduce that during the viral invasion and uncoating, M1249L complexed with vRNAP may serve as a temporary transcription factor to aid promoter binding and transcription. Although we purified ASFV vRNAP at different time points (8/12/16/24/48 hpi), no transcription factors were found to bind with vRNAP. Future studies would be necessary to reconstitute the endogenous vRNAP-transcription factor supercomplex by combining naturally extracted vRNAP with recombinant transcription/regulatory factors in vitro, which would shed light on the regulatory mechanisms of the complete vRNAP complex.
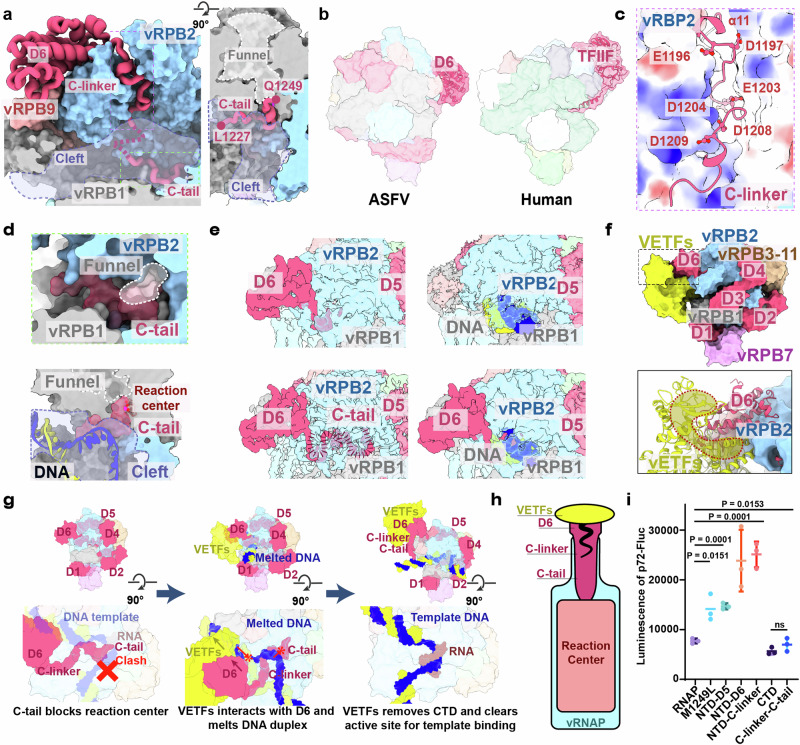
The CTD of M1249L consists of a compact α-helix structure (D6) followed by an unstructured C-linker and C-tail, with the middle amino acids 1219-1226 missing due to flexibility (Fig. 6a). The binding mode of M1249L CTD with the core vRNAP indicates that the sequential binding of D6, C-linker and C-tail is essential for the correct positioning of C-linker and C-tail into the reaction center and NTP channel (Fig. 6a, Supplementary Fig. 9). M1249L CTD is located on top of ‘lobe’ domain of vRPB2, and its α4-helix interacts with the N-terminal Zinc finger domain of vRPB9, which is absent in the core vRNAP (Supplementary Fig. 5). Coincidently, the binding position/mode of D6 is similar to that of human TFIIF (Fig. 6b)23, suggesting that D6 has similar functions to TFIIF in recruiting vRNAP to the promoter. The C-linker contains three pairs of acidic amino acids (E1196-D1197-E1203-D1204-D1208-D1209) that squeezes between ‘protrusion’ and ‘lobe’ domains of vRPB2, which are rich in positively charged residues (Fig. 6c). The terminal C-tail includes the last 23 residues including seven acidic amino acids, and extends across the reaction center and into the NTP channel (Fig. 6d). Dynamic maps show that when the C-tail is present at the reaction center, no nucleic acids are bound; conversely, when nucleic acids are bound at the reaction center, the C-tail is not observed, although the C-linker may still be present (Fig. 6e). This dynamic behavior is likely crucial for the switch on/off of vRNAP activity. In the model constructed based on the complete VACV vRNAP14, the CTD interacts with VETFs (Fig. 6f). Therefore, following promoter binding mediated by VETFl/s, the dissociation of VETFs from vRNAP would result in the detachment of the M1249L CTD, thereby pulling the C-linker and C-tail away from the reaction center, creating space for the initiation of mRNA synthesis (Fig. 6g). To investigate the regulatory mechanism of the M1249L CTD segment on vRNAP, we conducted cell-based functional assays, which demonstrated that co-expression of the CTD or C-linker&C-tail with vRNAP inhibits transcription, whereas deletion of the C-tail or C-linker&C-tail significantly increases the vRNAP activity. Interestingly, deletion of the entire CTD from M1249L showed no cumulative change, possibly due to the role of D6 in recruiting other VETF-like host factors (Fig. 6h and i).
Fig. 6. M1249L CTD is a molecular switch for vRNAP function.
a Location of M1249L CTD (in cartoon) in the vRNAP-M1249L complex (shown as surface). Cleft is highlighted by purple dash line. b Comparison of M1249L D6 domain (left) to human TFIIF (PDB: 7EG7) (right). c Acidic amino acids in M1249L C-linker region form a negatively charged cluster, interacting with nearby positively-charged vRNAP residues. d The perspective view shows the C-tail occupies NTP channel (left). The C-tail occupies the active center, preventing template entry and viral mRNA synthesis (right). e Four binding modes between M1249L CTD and vRNAP are identified: absence of C-tail when nucleic acids are absent, presence of whole CTD without nucleic acids, absence of CTD when nucleic acids are present, and absence of C-tail with presence of nucleic acids. f Modeling of ASFV vRNAP-M1249L-VETFs complex by superposing with VACV complete vRNAP (PDB: 7AMV). VETFs is colored yellow. Close-up view shows the interaction between M1249L-D6 and VETFs. g Schematic model of interaction between VETFs and D6, transforming vRNAP from a self-inhibitory state to an active state. Template DNA is in blue, complementary DNA in yellow, RNA in brown. “X” symbol represents steric clash between C-tail and DNA template. The asterisk arrow represents the progression direction. h Schematic diagram of VETFs binding to D6 and removing C-tail. The blue bottle represents vRNAP, the pink color represents reaction center, the magenta bottle cork represents M1249L CTD, and the yellow corkscrew represents VETFs. i HEK293T cells were transfected with plasmid of p72-Fluc and ASFV vRNAP with or without M1249L C terminal regions. Data are presented as mean values +/- SD (n = 3). Statistical analysis was performed by unpaired t test. P values are marked. “NTD-D5” includes RNAP and regions of M1249L from NTD to D5; “NTD-D6” includes RNAP and D6 of M1249L; “NTD-C-linker” includes RNAP and regions of M1249L from NTD to C-linker; “CTD” includes RNAP and CTD of M1249L; “C-linker-C-tail” includes RNAP and regions of M1249L from C-linker to C-tail.
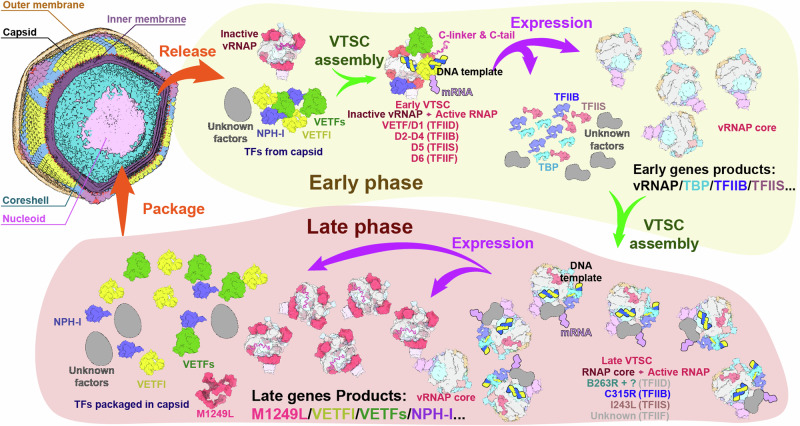
Based on previous proteomic characterization of purified ASFV viral particles and comprehensive bioinformatic analyses of the ASFV genome and transcriptome9, along with our structural and dynamic insights into the ASFV vRNAP and the M1249L protein complex, we herein propose a model for dynamic catalytic regulation mechanisms governing RNA transcription throughout the ASFV life cycle (Fig. 7). The initial step of viral infection starts with the virus entering the host cell. Upon invasion into host cell, ASFV undergoes uncoating and releases vRNAP (bound with M1249L) to initiate early transcription41. Accompanying the release of vRNAP, viral transcription factors such as VETFl/s, NPH-I, and CE, which are also part of the virions20, are recruited by the vRNAP-M1249L complex. These elements, together with the viral DNA genome, form the early pre-initiation complex (PIC). This newly formed PIC identifies the promoters of early transcription genes, triggering the early-phase transcription. At this stage, genes encoding vRNAP subunits and a secondary set of transcription factors including TBP, TFIIB, TFIIS, CE, and other unidentified factors, are transcribed, setting up the conditions for late-phase transcription. In the second step, the newly synthesized vRNAP and transcription factors produced during the early phase recognize and transcribe late viral genes by specific binding to the essential TATA-specific promoters42. The late genes include components of viral particles, as well as vRNAP subunits, M1249L, VETFl/s, NPH-1, CE, etc that are packaged into new virions. We speculate that the ASFV vRNAP-M1249L complex, in its inactivated state, would be incorporated into virions as a pre-assembled unit to promote the restart of early gene transcription in the subsequent infection cycle, a phenomenon observed in Arenavirus43 and possibly applicable to VACV16,44.
Fig. 7. Model of the ASFV transcription Dual Systems.
The core vRNAP and vRNAP-M1249L complex structures in this panel were derived from cryo-EM maps as depicted in Fig. 2b. The structural models of the other transcription factors are based on predictions generated by AlphaFold. Gray models signify unidentified proteins implicated in transcription. After the virus invades the host cell, the vRNAP-M1249L complex and VETFs, VETFl, and NPH-I transcription factors in the virons are released to initiate early transcription. Subsequently, the synthesis of vRNAP and virus-encoded transcription factors (TBP, TFIIB, TFIIS, CE, and other unknown factors) progresses, assembling the second form of RNA polymerase, setting the stage for the late phase transcription. In the late phase, newly synthesized vRNAP and transcription factors, produced during the early phase, initiate transcription of late viral genes by specifically binding to promoters of late phase genes including viral particle components including vRNAP, M1249L, VETFl/s, NPH-1, CE, etc., which are incorporated into new virions. The ASFV RNAP-M1249L complex in an inactivated state is included in virions as pre-assembled units to facilitate the resumption of early gene transcription in subsequent infection cycles.
Although structural and functional analyses have revealed the roles of M1249L in ASFV transcription, several important questions still remain unanswered. Does ASFV encode its own RNA polymerase I/III? If yes, which viral/host protein components are involved? Given that M1249L binds tightly with vRNAP, how is the binding/release of M1249L regulated during virus infection? The CE small subunit (D12) in VACV, which is a separately encoded protein, is fused to the C-terminal of vRPB7 in ASFV and therefore would restraint its conformational change. How does this fusion affect the assembly of CE on ASFV vRNAP during CCC formation? These questions are subjects for future research. On the other hand, in vitro assembly of ASFV vRNAP and transcription factors for structural and functional characterization could provide deeper insights into the regulatory mechanisms during ASFV infection. In addition, the N-terminal domain (NTD, residues 1-79) of Rap94 in VACV plays crucial roles in recruiting transcription factors and stabilizing the PIC, and may also function as a molecular switch for transcription. However, this region is missing in the solved structure of ASFV core vRNAP-M1249L complex, likely due to the absence of TFs that interact with and stabilize the NTD of M1249L. Future experiments aimed at obtaining the core vRNAP-M1249L-TFs complex would elucidate the role of M1249L NTD during ASFV transcription.
In conclusion, we present the structures of endogenous vRNAP of ASFV and its complexes with M1249L. Our findings reveal the multifunctional roles of M1249L as viral transcription factors involved in early transcription. These results highlight ASFV-specific structural features and pave the way to future control and treatment of ASFV.
Methods
Facility and ethics statements
The live viruses involved in this study were performed in the enhanced biosafety level 3 (P3 + ) facility of the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS), and approved by the Ministry of Agriculture and Rural Affairs. The protocols were approved by the Committee on the Ethics of Animal Experiments of the HVRI of CAAS (Approval numbers: 231017-01-GR). All animal-related research procedures complied with the Animal Welfare Act, Guide for the Care and Use of Laboratory Animals.
Cell and virus
Porcine alveolar macrophages (PAMs) and bone marrow cells (PBMs) were isolated from 5-week-old healthy specific pathogen free (SPF) piglets as described previously45,46 and maintained in RPMI-1640 medium (Gibco, USA) containing 20% porcine serum (HyClone, USA), 100 U/mL penicillin and streptomycin, and 2 mmol/mL glutamine at 37 °C with 5% CO2. HLJ/18-6GD was constructed and stored in HVRI as described previously19. ASFV Pig/Heilongjiang/2018 (HLJ/18) was isolated from field samples in China. PAM cells were infected with ASFV at a multiplicity of infection (MOI) of 0.5, and the supernatants were collected on day 3 post-infection. Viral titer was measured by using HAD assay as described previously46. To determine the dynamics of viral gene expression during ASFV replication in PAMs, the cells were infected with HLJ/18 at an MOI of 10. Then the cells were collected in TRIzol LS (Invitrogen, USA) for RNA extraction with 2 h interval from 0 h to 18 h post-infection.
RNA extraction, Library preparation, RNA sequencing, raw data processing, mapping and analysis
Total RNA was extracted by using TRIzil LS as the manufacturer described and quantified using Qubit 2.0 (Invitrogen, USA). The qualified RNA samples were added to magnetic beads (Illumina) with Oligo (dT) to enrich mRNA. Then the NEB Next® Ultra ™ RNA Library Prep Kit (NEW ENGLAND Biolabs) was used to construct the sequencing libraries. The AMPure XP beads were used to select the size of the fragments and purify the PCR products. Qualified libraries were pooled to the flowcell and sequenced by using Illumina Novaseq platform. The raw reads obtained by RNA sequencing were cleaned by using the Trim Galore (v0.6.5) to remove the adapter, and then were excluded low-quality reads (including reads with higher N content and reads with Q20-value ≤ 97% or Q30-value ≤ 93%)47 by using the fastp (v0.19.6). The filtered reads of RNA-seq data of ASFV-PAMs were aligned to the ASFV reference genome HLJ/18 (accession number: MK333180.1) by utilizing Hisat2 (v2.0.5)48. The SAM files generated by the above operation were sorted and converted into BAM files by using Samtools (v1.7)49, and featureCounts (v1.5.1)50 was further used to count the number of reads on each gene to generate counting matrices for subsequent analysis.
To further normalize the data mentioned above, we took the median of reads of all parallel data groups in a single period as the standard for normalization. We then normalized all data based on the proportions of the sum of data group in one period. Following that, we normalized the transcription quantity of each gene transcripts across all time periods, taking the lowest data point in the group as zero and the highest value as one to generate the final normalized data.
For each gene, the transcriptomic data obtained from independent measurements at different time points were compiled into multidimensional datasets. These datasets were then subjected to dimensionality reduction using the UMAP (Uniform Manifold Approximation and Projection) method. The resulting data points were grouped based on their expression peak times: transcripts peaking at or before 8 hours were categorized as immediate early transcripts, those peaking between 8 and 12 hours as early transcripts, and those peaking after 12 hours as late transcripts. Transcripts without a clear peak in these time frames were classified as immediate transcripts.
Construction of Twin strep-tagged recombinant ASFV
The recombinant ASFV tagged with Twin strep (Abbreviated ASFV-1242TS) was constructed from the attenuated ASF vaccine candidate HLJ/18-6GD with a twin-strep tag fused at the end of EP1242L gene by using homologous recombination as described previously19. To facilitate screening the recombinant virus, mCherry gene driven by ASFV p72 promoter was cis-inserted before EP1242L gene. Recombinant transfer vectors containing the flanking genomic regions of EP1242L gene, mapping approximately 700 bp to the left and right of the inserted parts, and the reporter gene cassettes containing the reporter gene of mCherry with the ASFV p72 late gene promoter were used as shown in Fig. 1. To make sure the correct termination of EP1242L fusing twin-strep, the sequence of 40 bp trans-following the original EP1242L gene ORF retains between mCherry ORF and EP1242L fusing twin-strep. Recombinant transfer vector was constructed by using fusing PCR and the Gibson Assembly technique (Invitrogen Life Sciences, USA). PAMs were transfected with the constructed transfer vectors by using the TransIT LT1 transfection reagent (Mirus Bio, Madison, WI, USA) and then infected with HLJ/18-6GD at 24 h post-transfection. The recombinant virus was purified by successively picking fluorescent plaques combined with limited dilution in PAMs. The purified recombinant ASFV-1242TS was amplified in PBMs and confirmed by sequencing.
Virus growth detection
The growth kinetics of HLJ/18-6GD and recombinant ASFV-1242TS were assessed in PAMs. PAMs were infected with ASFVs at an MOI of 0.1, and cell supernatants were collected at 0, 24, 48, 72, 96, 120, and 144 h post-infection. Viral genomic DNA in the supernatants was extracted and detected for viral p72 gene copy numbers by using qPCR recommended by WOAH51.
Purification of ASFV RNAP
For purification of ASFV vRNAP from infected cells, PBM cells were grown in cell factory up to 80%-90% of confluence. The cells were infected with purified ASFV-1242TS virus with a MOI of 1. After 48 hours, the cells were centrifuged to collect the cell pellet, which was then resuspended in PBS. Following another centrifugation, the supernatant was discarded and the cell pellet was frozen at -80 °C for storage. Cell pellets were resuspended in buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 1‰ Tween-20) supplemented with a proteases inhibitor (cOmplete EDTA-free Protease Inhibitor Cocktail Tablet, Roche) and lysed with a Dounce homogenizer on ice (40 plunges). Cell lysate was centrifuged at 3000 g for 50 min at 4 °C, then the extract was incubated with 0.5 ml Strep-Tactin XT beads for 2 hours at 4 °C. The beads were collected by centrifugation at 500 g for 3 min at 4 °C and the target protein was eluted using an elution buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 1.5 mM MgCl2, 1 mM DTT 50 mM biotin) for three times. The purified protein complex was flash-frozen in liquid nitrogen and stored at −80 °C until use.
Mass photometry analysis of vRNAP complex
Mass photometry (MP) is a light-scattering-based technique that measures the mass of individual molecules to obtain molecular mass distributions of proteins and complexes in solution. Mass photometry was conducted using Refeyn OneMP (Refeyn Ltd) with data acquisition facilitated by AcquireMP software (Refeyn Ltd v.2.3). Microscope coverslips (Corning, 24 × 50 mm, thickness 1.5) were prepared by washing with ddH2O and isopropanol, followed by the placement of silicone templates to create reaction chambers prior to measurement. Calibration of the instrument was performed using BSA dissolved in 1xPBS. Purified vRNAP samples and buffer (as above) were filtered by 0.22 μm membrane prior to Mass photometry analysis. A volume of 8 μl protein buffer was added to each well, and the focal position was determined and fixed. Subsequently, 1 μl of protein solution (to a final concentration of ~100 nM) was added to the well and mixed thoroughly. Mass photometry signals were recorded for 60 s to ensure the detection of ~1000 individual binding events. Data analysis was conducted using DiscoverMP software.
Cryo-EM sample preparation and data collection
All cryo-EM samples preparation underwent a standardized preprocessing pipeline. To prepare cryo-EM samples, 4 μl of purified ASFV RNA-Pol complex in a buffer containing 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 0.5 mM BS3 was applied to each grid. Amorphous alloy films (R1.2/1.3, Au, 300 mesh) were glow-discharged for 90 seconds prior to the application of the complex at 0.8 mg/ml concentration. The grids were blotted at 22 °C for 7 s at ~100% humidity and plunged into liquid ethane using an FEI Vitrobot Mark IV. The samples were loaded onto the FEI Titan Krios transmission electron microscope at 300 kV with a GIF-Quantum energy filter (Gatan) and a Gatan K2-summit detector for data collection. Automatic data collection used SerialEM software (http://bio3d.colorado.edu/SerialEM/). The nominal magnification of ×96,000 corresponds to a calibrated pixel size of 0.81 Å at the specimen and a dose rate of 10 e-/pixel/s. Each image was exposed for 6.5 s to obtain an accumulative dose of ~40 e-/Å2, fractioned into 32 frames. The final defocus range of all images sets was approximately –1.5 to –2.2 μm.
Image processing
All movies selected for this analysis underwent a standardized preprocessing pipeline. Beam-induced motion was corrected, and frames were summed for each movie using MotionCorr252 with a 5 × 5 patch strategy. The contrast transfer function (CTF) parameters for each summed image were estimated using the Patch CTF module in cryoSPARC53. Subsequently, particles were picked and extracted from the dose-weighted images employing the cryoSPARC Blob Picker and Particle Extraction tools.
For datasets collected of 48 hr (h.p.i. = 48), the complete dataset comprised 12,224 motion-corrected images, yielding 4,417,795 particles. Using cryoSPARC 2D classification (n = 200), 1,025,118 particles were identified as high-quality. Initial 3D maps were generated from a subset of these particles using cryoSPARC Ab initio Reconstruction. Further refinement was performed using cryoSPARC Heterogeneous Refinement (n = 5) and generated Class 2 and Class 3 containing 472,243 and 351,835 particles as high-quality, respectively. These were subsequently refined via cryoSPARC Homogeneous Refinement, resulting in two high-resolution maps at 2.56 Å (state 1) and 2.67 Å (state 2).
To elucidate the structures containing the D1 domain of M1249L, a local 3D classification was employed to isolate particles incorporating D1 using RELION54 3D Classification. Following homogeneous and local refinement in cryoSPARC, two maps at resolutions of 2.64 Å (state 3) and 2.97 Å (state 4) were obtained. A further local 3D classification focused on the D2 domain of M1249L led to the selection of 61,437 particles for the reconstruction of the D2-containing complex (state 5) and refined to a resolution of 2.82 Å. All 61,437 particles underwent a third round of local 3D classification targeting the region between the D2 and D4 domains of M1249L, from which 10,433 particles were chosen to reconstruct the complete M1249L-vRNAP structure (state 5) at a resolution of 3.68 Å (see Supplementary Fig. 2).
To capture all states of M1249L, datasets from 12 hr (7645 movies), 16 hr (7650 movies), and 24 hr (9587 movies) intervals were processed, extracting 1,115,057, 1,565,450, and 1,648,320 particles, respectively (see Supplementary Fig. 4). 2D Classification, ab initio Reconstruction, and Heterogeneous Refinement imposed in cryoSPARC were employed to discard poor-quality particles from each dataset. To avoid bias from averaging different states and to identify period-specific states, particles from distinct collection periods were processed separately. This involved four rounds of 3D classification, ensuring each final class contained fewer than 10,000 particles. The initial 3D classification aimed to segregate particles based on the presence of nucleic acid within the vRNAP cleft. Subsequently, three rounds of template-free 3D classification generated 250 classes from each dataset of various time periods. Particles from different datasets displaying same features were manually merged based on 3D classification results, yielding 25 distinct states of the complex. These groups of particles were then refined using RELION’s Refine3D. All resolutions and particle counts for the final reconstructions detailed are shown in Supplementary Table 1.
All reported resolutions are based on the gold-standard Fourier shell correction (FSC) = 0.143 criterion. RESMAP55 is used to estimate local resolution.
Model building and refinement
The subunits in the RNA polymerase were each identified by Peptide Mass Fingerprinting with MALDI-TOF-MS from the Institute of Biophysics. To build and refine our models, the program AlphaFold56 was used to predict the corresponding subunits of ASFV RNA polymerase. Program UCSF chimera57 was used to position the coordinates of the model into the 2.56 Å map. Further optimization of the positioning of the side chains were performed using the program COOT58 and the real-space refinement was performed using the program PHENIX59. The core RNA polymerase structure was modeled to the 2.67 Å map and refined similar to the full RNA polymerase.
Phylogenetic analysis of RNA polymerase from different species
Sequences of RPB1 and RPB2 from swine (RPB1:UniProt:A0A7M4DUC2; RPB2:UniProt:I3LGP4), yeast (RPB1:UNIPROT:P04050; RPB2:UNIPROT:P08518), ASFV (vRPB1:GenBank: QBH90587.1; vRPB2 GenBank: QBH90541.1), and VACV (Rpo147-RPB1:PDB:6RFL_A; Rpo132-RPB2:PDB:6RFL_B) were obtained from UniProt. These sequences were aligned using ClustalW, and a phylogenetic tree was constructed with the Maximum Likelihood method in MEGA, then visualized with EvolView60.
ASFV vRNAP activity detection in vitro
HEK293T cells were transfected with plasmids of p72-Fluc RNAP reporter system in 24-well plates as described previously21. Specifically, the eight vRNAP subunits and M1249L are each encoded by an individual EF1α- Flag lentiviral plasmid via the Gateway cloning system (Invitrogen, USA, 11789021), together with the plasmid encoding p72-Fluc. The amounts of each plasmid were shown below: p72-Fluc 400 ng, C315R 300 ng, D205R 50 ng, EP1242L 75 ng, C147L 50 ng, CP80R 50 ng, NP1450L 75 ng, H359L 50 ng, C122R 50 ng, with or without M1249L. At 36 h post-incubation, the activities of firefly luciferase were measured by using the Luciferase Reporter Assay System (Promega Corporation, E1501) according to the manufacturer’s instructions. Data were analyzed by using GraphPad Prism.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We particularly thank Prof. Jingfei Wang from Harbin Veterinary Research Institute, the Chinese Academy of Agricultural Sciences for the technical assistance during sample preparation. We thank Yan Wu from Institute of Biophysics for his research assistant service. All research described in this article is supported by National Key Research and Development Program (2018YFA0900801, 2023YFC2306603), Chinese Academy of Sciences (YSBR-010, XDB37030200), National Science Foundation(12034006, T2394482, 32200135), National Science Fund for Distinguished Young Scholar (32325004), NSFC Innovative Research Group (81921005), NSFC General Program (32270179), the Heilongjiang Provincial Natural Science Foundation of China (JQ2023C005), Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-CSLPDCP-202301), and the Central Public-interest Scientific Institution Basal Research Fund (CAAS-ZDRW202409).
Author contributions
X.W., D.Z., Z.R., and Z.B. conceived the study. Z.Z., R.Z., contributed to molecular cloning, cell culture, and protein expression; X.F., K.X., T.Z., Y.Y., and Y.C. contributed to protein purification; X.F., Z.Z., X.L., and X.O. performed the in vitro assays’ experiments; N.W., X.F., and Y.C. carried out the structural studies by cryo-EM; D.Z., N.W., X.F., Z.Z., Y.C. and X.W. contributed to manuscript writing; N.W., X.F., and Y.C. contributed to figure preparation.
Peer review
Peer review information
Nature Communications thanks Wim Burmeister, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The cryo-EM maps and coordinates have been deposited to the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB). The transcribing complexes include ASFV RNA polymerase core (EMD-39507, PDB ID 8YQV), ASFV RNA polymerase-M1249L complex1 (EMD-39506, PDB ID 8YQU), ASFV RNA polymerase-M1249L complex2 (EMD-39505, PDB ID 8YQT), ASFV RNA polymerase-M1249L complex3 (EMD-39508, PDB ID 8YQW), ASFV RNA polymerase-M1249L complex4 (EMD-39509, PDB ID 8YQX), ASFV RNA polymerase-M1249L complex complete (EMD-39510, PDB ID 8YQY), ASFV RNA polymerase-M1249L-DNA complex (EMD-39511, PDB ID 8YQZ), ASFV RNA polymerase-M1249L complex5 (EMD-39514), ASFV RNA polymerase-M1249L complex6 (EMD-39512), ASFV RNA polymerase-M1249L complex7 (EMD-39513), ASFV RNA polymerase-M1249L complex8 (EMD-39516), ASFV RNA polymerase-M1249L complex9 (EMD-39515), ASFV RNA polymerase-M1249L complex10 (EMD-39517), ASFV RNA polymerase-M1249L complex11 (EMD-39518), ASFV RNA polymerase-M1249L complex12 (EMD-39519), ASFV RNA polymerase-M1249L complex13 (EMD-39521), ASFV RNA polymerase-M1249L complex14 (EMD-39520), ASFV RNA polymerase-M1249L complex15 (EMD-39522), ASFV RNA polymerase-M1249L complex16 (EMD-39523), ASFV RNA polymerase-M1249L complex17 (EMD-39524), ASFV RNA polymerase-M1249L complex18 (EMD-39525), ASFV RNA polymerase-M1249L complex19 (EMD-39526), ASFV RNA polymerase-M1249L complex20 (EMD-39527), ASFV RNA polymerase-M1249L complex21 (EMD-39528), ASFV RNA polymerase-M1249L complex22 (EMD-39530), ASFV RNA polymerase-M1249L complex23 (EMD-39531), ASFV RNA polymerase-M1249L complex24 (EMD-39529), and ASFV RNA polymerase-M1249L complex25 (EMD-39536). The raw sequencing data reported in this paper were submitted to Genome Sequence Archive (GSA) of National Genomics Data Center (http://bigd.big.ac.cn, Bioproject number: PRJCA003613). Other source data are also provided with this paper. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dongming Zhao, Nan Wang, Xiaoying Feng, Zhenjiang Zhang.
Contributor Information
Zihe Rao, Email: raozh@ibp.ac.cn.
Zhigao Bu, Email: buzhigao@caas.cn.
Yutao Chen, Email: chenyutao@ibp.ac.cn.
Xiangxi Wang, Email: xiangxi@ibp.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54461-1.
References
- 1.Gaudreault, N. N., Madden, D. W., Wilson, W. C., Trujillo, J. D. & Richt, J. A. African Swine Fever Virus: An Emerging DNA Arbovirus. Front. Vet. Sci.710.3389/Fvets.2020.00215 (2020). [DOI] [PMC free article] [PubMed]
- 2.Salas, M. L. & Andrés, G. African swine fever virus morphogenesis. Virus Res173, 29–41 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Wang, N. et al. Architecture of African swine fever virus and implications for viral assembly. Science366, 640–644 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Hernaez, B. & Alonso, C. Dynamin- and clathrin-dependent endocytosis in african swine fever virus entry. J. Virol.84, 2100–2109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonso, C. et al. African swine fever virus-cell interactions: from virus entry to cell survival. Virus Res173, 42–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballester, M. et al. Disruption of nuclear organization during the initial phase of african swine fever virus infection. J. Virol.85, 8263–8269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortin, J. & Vinuela, E. Requirement of cell-nucleus for african swine fever virus-replication in vero cells. J. Virol.21, 902–905 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simoes, M., Freitas, F. B., Leitao, A., Martins, C. & Ferreira, F. African swine fever virus replication events and cell nucleus: New insights and perspectives. Virus Res27010.1016/j.virusres.2019.197667 (2019). [DOI] [PubMed]
- 9.Cackett, G. et al. The African swine fever virus transcriptome. J Virol9410.1128/JVI.00119-20 (2020). [DOI] [PMC free article] [PubMed]
- 10.Dixon, L. K., Chapman, D. A. G., Netherton, C. L. & Upton, C. African swine fever virus replication and genomics. Virus Res173, 3–14 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez, J. M. & Salas, M. L. African swine fever virus transcription. Virus Res173, 15–28 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Kuznar, J., Salas, M. L. & Vinuela, E. Rnas synthesized invitro by purified african swine fever virus. Arch. Biol. Med Exp.13, 465–465 (1980). [Google Scholar]
- 13.Muñoz-Moreno, R., Galindo, I., Cuesta-Geijo, M. A., Barrado-Gil, L. & Alonso, C. Host cell targets for African swine fever virus. Virus Res209, 118–127 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Grimm, C., Bartuli, J., Boettcher, B., Szalay, A. A. & Fischer, U. Structural basis of the complete poxvirus transcription initiation process. Nat. Struct. Mol. Biol.28, 779–788 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Hillen, H. S. et al. Structural basis of poxvirus transcription: transcribing and capping vaccinia complexes. Cell179, 1525–1536 e1512 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Grimm, C. et al. Structural basis of poxvirus transcription: vaccinia rna polymerase complexes. Cell179, 1537–1550 e1519 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Cackett, G., Sykora, M. & Werner, F. Transcriptome view of a killer: African swine fever virus. Biochem Soc. Trans.48, 1569–1581 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilotto, S., Sýkora, M., Cackett, G., Dulson, C. & Werner, F. Structure of the recombinant RNA polymerase from African Swine Fever Virus. Nat. Commun.15, 1606 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, W. Y. et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci.63, 623–634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alejo, A., Matamoros, T., Guerra, M. & Andres, G. A proteomic atlas of the african swine fever virus particle. J. Virol.92 (2018). [DOI] [PMC free article] [PubMed]
- 21.Zhang, Y. H. et al. ASFV transcription reporter screening system identifies ailanthone as a broad antiviral compound. Virologica Sin.38, 459–469 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armache, K. J., Mitterweger, S., Meinhart, A. & Cramer, P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem.280, 7131–7134 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Chen, X. Z. et al. Structural insights into preinitiation complex assembly on core promoters. Science372, 480 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Kettenberger, H., Armache, K. J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell16, 955–965 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res50, W210–W215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madej, T. et al. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res42, D297–D303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madej, T., Marchler-Bauer, A., Lanczycki, C., Zhang, D. & Bryant, S. H. in Structural Bioinformatics: Methods and Protocols (ed Zoltán Gáspári) 175–186 (Springer US, 2020).
- 28.Cramer, P. et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science288, 640–649 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Nikolov, D. B. et al. Crystal-structure of a Tfiib-Tbp-Tata-element ternary complex. Nature377, 119–128 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Chen, H. T. & Hahn, S. Binding of TFIIB to RNA polymerase II: Mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell12, 437–447 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Liu, X., Bushnell, D. A., Wang, D., Calero, G. & Kornberg, R. D. Structure of an RNA Polymerase II-TFIIB Complex and the Transcription Initiation Mechanism. Science327, 206–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kostrewa, D. et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature462, 323–330 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Sainsbury, S., Niesser, J. & Cramer, P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature493, 437–U191 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Bushnell, D. A., Westover, K. D., Davis, R. E. & Kornberg, R. D. Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science303, 983–988 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Farnung, L., Ochmann, M., Garg, G., Vos, S. M. & Cramer, P. Structure of a backtracked hexasomal intermediate of nucleosome transcription. Mol. Cell82, 3126 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Jeon, C. J., Yoon, H. S. & Agarwal, K. The transcription factor tfiis zinc ribbon dipeptide asp-glu is critical for stimulation of elongation and rna cleavage by Rna-Polymerase-Ii. Proc. Natl Acad. Sci. USA91, 9106–9110 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kettenberger, H., Armache, K. J. & Cramer, P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell114, 347–357 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Liu, H. & Cheng, L. Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science349, 1347–1350 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Tng, P. Y. L., Al-Adwani, L., Pauletto, E., Hui, J. Y. K. & Netherton, C. L. Capsid-specific antibody responses of domestic pigs immunized with low-virulent african swine fever virus. Vaccines-Basel1110.3390/Vaccines11101577 (2023). [DOI] [PMC free article] [PubMed]
- 40.Cui, S. et al. African swine fever virus M1249L protein antagonizes type I interferon production via suppressing phosphorylation of TBK1 and degrading IRF3. Virus Res319, 1872–7492 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Yáñez, R. J. et al. African swine fever virus encodes two genes which share significant homology with the two largest subunits of DNA-dependent RNA polymerases. Nucleic Acids Res21, 2423–2427 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.García-Escudero, R. & Viñuela, E. Structure of African swine fever virus late promoters:: Requirement of a TATA sequence at the initiation region. J. Virol.74, 8176–8182 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang, H. L. et al. Structural basis for recognition and regulation of arenavirus polymerase L by Z protein. Nat Commun1210.1038/S41467-021-24458-1 (2021). [DOI] [PMC free article] [PubMed]
- 44.Davison, A. J. & Moss, B. Structure of vaccinia virus early promoters. J. Mol. Biol.210, 749–769 (1989). [DOI] [PubMed] [Google Scholar]
- 45.Rathakrishnan, A., Reis, A. L., Moffat, K. & Dixon, L. K. Isolation of porcine bone marrow cells and generation of recombinant african swine fever viruses. Methods Mol. Biol.2503, 73–94 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Zhao, D. et al. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes Infect.8, 438–447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, H. et al. The Sequence Alignment/Map format and SAMtools. BIOINFORMATICS25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 51.2019, O. T. M. African Swine Fever. OIE Terrestrial ManualChapter 3.8.1 (2019).
- 52.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Scheres, S. H. W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol.180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swint-Kruse, L. & Brown, C. S. Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics21, 3327–3328 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. computational Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D., Biol. Crystallogr.60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D., Biol. Crystallogr.66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian, B., Gao, S., Lercher, M. J., Hu, S. & Chen, W.-H. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res47, W270–W275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps and coordinates have been deposited to the Electron Microscopy Data Bank (EMDB) and Protein Data Bank (PDB). The transcribing complexes include ASFV RNA polymerase core (EMD-39507, PDB ID 8YQV), ASFV RNA polymerase-M1249L complex1 (EMD-39506, PDB ID 8YQU), ASFV RNA polymerase-M1249L complex2 (EMD-39505, PDB ID 8YQT), ASFV RNA polymerase-M1249L complex3 (EMD-39508, PDB ID 8YQW), ASFV RNA polymerase-M1249L complex4 (EMD-39509, PDB ID 8YQX), ASFV RNA polymerase-M1249L complex complete (EMD-39510, PDB ID 8YQY), ASFV RNA polymerase-M1249L-DNA complex (EMD-39511, PDB ID 8YQZ), ASFV RNA polymerase-M1249L complex5 (EMD-39514), ASFV RNA polymerase-M1249L complex6 (EMD-39512), ASFV RNA polymerase-M1249L complex7 (EMD-39513), ASFV RNA polymerase-M1249L complex8 (EMD-39516), ASFV RNA polymerase-M1249L complex9 (EMD-39515), ASFV RNA polymerase-M1249L complex10 (EMD-39517), ASFV RNA polymerase-M1249L complex11 (EMD-39518), ASFV RNA polymerase-M1249L complex12 (EMD-39519), ASFV RNA polymerase-M1249L complex13 (EMD-39521), ASFV RNA polymerase-M1249L complex14 (EMD-39520), ASFV RNA polymerase-M1249L complex15 (EMD-39522), ASFV RNA polymerase-M1249L complex16 (EMD-39523), ASFV RNA polymerase-M1249L complex17 (EMD-39524), ASFV RNA polymerase-M1249L complex18 (EMD-39525), ASFV RNA polymerase-M1249L complex19 (EMD-39526), ASFV RNA polymerase-M1249L complex20 (EMD-39527), ASFV RNA polymerase-M1249L complex21 (EMD-39528), ASFV RNA polymerase-M1249L complex22 (EMD-39530), ASFV RNA polymerase-M1249L complex23 (EMD-39531), ASFV RNA polymerase-M1249L complex24 (EMD-39529), and ASFV RNA polymerase-M1249L complex25 (EMD-39536). The raw sequencing data reported in this paper were submitted to Genome Sequence Archive (GSA) of National Genomics Data Center (http://bigd.big.ac.cn, Bioproject number: PRJCA003613). Other source data are also provided with this paper. Source data are provided with this paper.