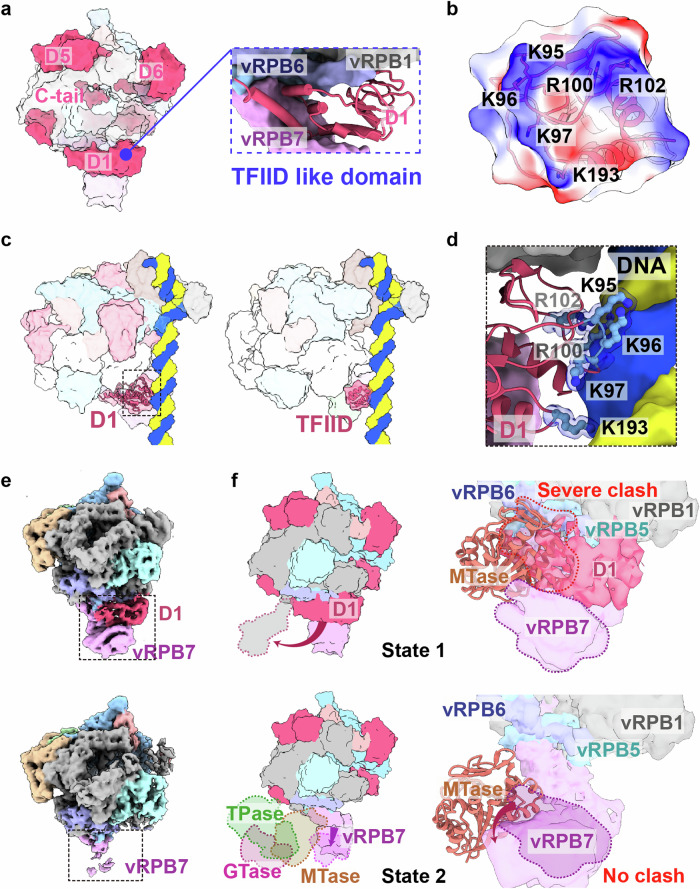
Fig. 3. Structural analysis of the M1249L D1 domain reveals its role as a transient transcription factor similar to TFIID.
a The surface structure of vRNAP-M1249L, highlighting the functional domains of M1249L, shows similarities to the classical transcription factor TFIID (see blue dashed box). b Surface potential map of M1249L D1 domain in contact with nucleic acids. c Left: ASFV RNAP-M1249L complex superposed onto Sus scrofa Pol II (PDB: 7EG7). Right: Sus scrofa Pol II (PDB: 7EG7). M1249L D1 is presented in cartoon format, with black dashed lines indicating the positions where it contacts the nucleic acid. d Details of key amino acids involved in M1249L D1-nucleic acid interactions. M1249L D1 domain depicted in cartoon, with crucial amino acids shown in ball-and-stick mode. e Two different cryo-EM maps demonstrate the stabilizing effect of M1249L D1 domain on vRPB7. f The two states of M1249L D1 domain when CE is unbound (top) or bound (bottom) to vRNAP are shown in the surface. The binding state of CE is obtained with reference to VACV CCC complex (PDB: 6RIE). The three domains of CE are colored respectively as green (TPase), magenta (GTase), and orange (MTase). The zoom-in image shows a significant clash between D1 and MTase when D1 is bound to vRNAP (top), while there is no clash when D1 moves away (bottom). The structure of the ASFV MTase is predicted by AlphaFold, displayed in cartoon format, with vRNAP shown as a surface representation.