Abstract
Studies of laminopathy-based progeria offer insights into aging-associated diseases and highlight the role of LMNA in chromatin organization. Mandibuloacral dysplasia type A (MAD) is a largely unexplored form of atypical progeria that lacks lamin A post-translational processing defects. Using iPSCs derived from a male MAD patient carrying homozygous LMNA p.R527C, premature aging phenotypes are recapitulated in multiple mesenchymal lineages, including mesenchymal stem cells (MSCs). Comparison with 26 human aging MSC expression datasets reveals that MAD-MSCs exhibit the highest similarity to senescent primary human MSCs. Lamina-chromatin interaction analysis reveals reorganization of lamina-associating domains (LADs) and repositioning of non-LAD binding peaks may contribute to the observed accelerated senescence. Additionally, 3D genome organization further supports hierarchical chromatin disorganization in MAD stem cells, alongside dysregulation of genes involved in epigenetic modification, stem cell fate maintenance, senescence, and geroprotection. Together, these findings suggest LMNA missense mutation is linked to chromatin alterations in an atypical progeroid syndrome.
Subject terms: Mesenchymal stem cells, Senescence, Epigenomics
Atypical progeria syndromes provide insights into aging and chromatin organization. Here, authors show that a LMNA mutation in mandibuloacral dysplasia disrupt chromatin structure and accelerate senescence in mesenchymal stem cells.
Introduction
Nuclear lamins are ubiquitously expressed type V intermediate filament proteins that are polymerized and assembled, with other interacting proteins, into nuclear lamina, a structure essential for nuclear integrity and chromatin organization. Mutations in the LMNA give rise to a variety of disorders, ranging from muscular dystrophy, lipodystrophy, cardiomyopathy, and neuropathy to severe progeroid syndromes, which are collectively referred to as laminopathies1. In addition to the extensively studied typical laminopathy-based progeroid syndrome (TPS), e.g., Hutchinson-Gilford progeria syndrome (HGPS), a number of LMNA variants have been reported to cause atypical progeroid syndrome (APS)2–5. Mandibuloacral dysplasia type A (MAD), a rare genetic disorder resulting from LMNA recessive mutation, is characterized by mandibular hypoplasia, cutaneous atrophy, acro-osteolysis, lipodystrophy, growth retardation, and premature death and represents a classical APS6. However, little is known about the molecular changes and the underlying mechanism behind MAD pathogenesis.
The difference between TPS and APS stems primarily from the production and accumulation of progerin or prelamin A, which competes with lamin A/C for the interaction with lamina-associated proteins, including DNA damage repair-associated proteins (DNAPKcs7, PARP18, TRF29, SIRT110, and SIRT611) and epigenetic modifiers (RBBP4/712, SUV39H113, HDAC214 and Polycomb proteins15,16), while specifically impairing the mitochondrial fitness-associated PGC1α17 and anti-oxidation related NRF218. These findings serve as, at least partially, the driving force behind HGPS pathogenesis, while the LMNA variants presented in APS indicate a different mechanism than TPS. Therefore, to understand how the specific LMNA mutation drives the pathogenesis in MAD, it is necessary to examine the fundamental functions of lamin A/C in chromatin organization.
Apart from as a nuclear structural component, lamins closely interact with chromatin to form lamina-associating domains (LADs) at the nuclear periphery19. Loss or gain of these critical contacts leads to gene dysregulation and links to the pathogenesis of multiple laminopathies and the aging process20–23. Lamina-chromatin interaction also facilitates higher-order chromatin structure organization24. Indeed, loss of nuclear integrity is frequently observed in laminopathies, which likely mirrors chromatin disorganizations. Although individual alterations of the chromatin, such as accessibility25,26, enhancer remodeling27–29, histone modifications30,31, topologically associating domains (TADs)32, and chromatin compartmentalization23,33–35, have been investigated in senescence or aging, few of these have been explored in MAD or other APS.
In this study, iPSCs derived from a male MAD patient harboring the LMNA p.R527C mutation demonstrated features of premature aging. Upon induced differentiation, these cells exhibited accelerated senescence in mesenchymal lineages. Integrative analysis of aging hMSC datasets with those from MAD-MSCs identified gene expression profiles associated with geroprotection and senescence, revealing a high degree of similarity between MAD-MSCs and senescent primary human MSCs(hMSCs). Multi-omic profiling of chromatin features identified significant alterations in the chromatin architecture of MAD-MSCs, which were linked to the dysregulation of key genes involved in epigenetic modifications, lineage specification, senescence, and geroprotection. Considering the similarities in molecular alterations and clinical manifestations between APS and physiological aging, this study not only provides insights into normal aging and aging-related degenerations but also suggests potential mechanisms of chromatin disorganization for other laminopathies.
Results
iPSCs-derived mesenchymal lineages from a MAD patient exhibited accelerated senescence
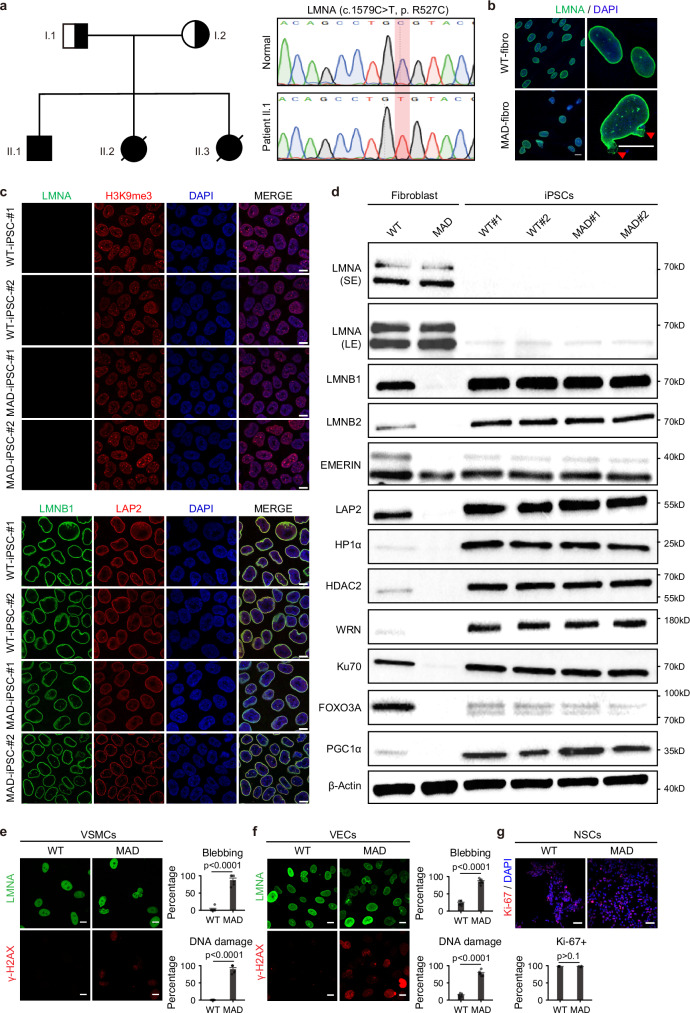
A previously reported male MAD patient was confirmed to carry the c.1579 C > T, p. R527C mutation in the LMNA gene (Fig. 1a). Isolated patient fibroblasts did not show the additional visible isoform of lamin A, though mutant cells exhibited earlier accelerated senescence, higher percentage of misshapen nuclear morphology (69.6 ± 9.3 % in MAD vs 10.8 ± 5.7 % in control, p = 0.0004) and abnormal epigenetic modifications (Fig. 1a and Supplementary Fig. 1a–j). The patient-specific iPSCs were generated using minicircle DNA as described previously36, and two independent clones were picked out for further validation (denoted as MAD-iPSCs #1 and #2). The two WT-iPSCs clones were generated from a single healthy individual as previously described36, and the WT-iPSCs and respective derivatives from WT-iPSCs were used as control throughout the study. No random integration of the minicircle vector was detected in these iPSCs (Supplementary Fig. 2a). Reverse transcription PCR and quantitative PCR (qPCR) showed comparable expression levels of pluripotency-associated genes in iPSCs and human ESCs H9 (Supplementary Fig. 2b, c). The bisulfite sequencing of endogenous OCT3/4 promoter region demonstrated rewriting of the DNA methylation after reprogramming (Supplementary Fig. 2d). The pluripotency and differentiation potential of these iPSCs were further confirmed by immunostaining and in vivo teratoma formation assay (Supplementary Fig. 2e, f). Karyotyping analysis also ensured the genome integrity in the iPSCs (Supplementary Fig. 2g).
Fig. 1. Mesenchymal lineages derived from MAD iPSCs manifested progeroid defects.
a The pedigree of a MAD family and Sanger sequencing confirming the homozygous point mutation in LMNA (c.1579 C > T, p. R527C). b Immunostaining of lamin A/C (LMNA, green) in normal and MAD-patient derived dermal fibroblasts. DAPI, blue. Scale bar 10μm. The experiment was repeated three times with similar results. c Immunostaining of lamin A/C (LMNA, green), H3K9me3 (red), lamin B1 (LMNB1, green) and LAP2 (red) in WT-iPSCs and MAD iPSCs clones. DAPI, blue. Scale bar 10μm. The experiment was repeated three times with similar results. d Immunoblotting of lamin A/C (LMNA), lamin B1 (LMNB1), lamin B2 (LMNB2), EMERIN, LAP2, and WRN, KU70; HADC2, HP1a and FOXO3, PGC1a in dermal fibroblasts and two independent WT- and MAD- iPSCs clones. β-Actin was used as the loading control and only shown once as limited space. The experiment was repeated three times with similar results. e Representative images of lamin A/C (LMNA, green) and γ-H2A.X (red) co-immunostaining in passage 8 (P8) of WT and MAD-VSMCs with a corresponding statistical analysis. DAPI, blue. Scale bar 10 μm. Data are mean ± SD, the p value was calculated using two-tailed unpaired t-test, n = 6. Three independent differentiation experiments were performed with similar results. f Representative images of lamin A/C (LMNA, green) and γ-H2A.X (red) co-immunostaining in passage 12 (P12) of WT and MAD-VECs with a corresponding statistical analysis. DAPI, blue. Scale bar 10 μm. Data are mean ± SD, the p value was calculated using two-tailed unpaired t-test, n = 7. Three independentdifferentiation experiments were performed with similar results. g Representative images of Ki 67 (red) staining in WT-NSCs and MAD-NSCs at passage 20. Data are mean ± SD, the p value was calculated using two-tailed unpaired t-test, n = 4. Three independent differentiation experiments were performed with similar results. DAPI, blue. Scale bar 10 μm. Source data are provided as a Source Data file.
Next, we examined the nuclear integrity and aging-associated phenotypes in iPSCs. Lamin A/C was barely detectable in MAD-iPSCs, whereas lamin B1 was restored (Fig. 1c, d and Supplementary Fig. 3b). The decreased expression of lamin B2, LAP2, WRN, Ku70, FOXO3a, PGC1α, HP1α, and HDAC2 were all restored in MAD-iPSCs (Fig. 1d and Supplementary Fig. 3a). Furthermore, nuclear dysmorphology was rescued and proliferative capability was restored in MAD-iPSCs (Supplementary Fig. 3a and 1i). These data collectively indicated that premature senescence in MAD fibroblasts was rejuvenated in the pluripotent state.
To recapitulate the tissue-specific defects of MAD, we differentiated MAD-iPSCs into multiple lineages, including vascular smooth muscle cells (VSMCs), vascular endothelial cells (VECs), and neural stem cells (NSCs). The differentiated MAD-VSMCs, marked by Calponin, smooth muscle alpha-actin (αSMA), and smooth muscle 22α (SM22α) (Supplementary Fig. 4d–f, j), exhibited a remarkable increase in nuclear deformation (90.6 ± 13.7% in MAD-VSMCs vs 2.0 ± 5.9% in controls; p < 0.0001) and elevated DNA damage at passage 5 (90.0 ± 10.9 % in MAD-VSMCs vs not detectable in controls; p < 0.0001) (Fig. 1e). MAD-VECs, marked by von Willebrand factor (VWF) and endothelial nitric oxide synthase (eNOS) and validated by the Ac-LDL up-taking function (Supplementary Fig. 4g–i, j), manifested significantly elevated frequencies of nuclear blebbing (78.3 ± 7.2% in MAD-VECs vs 12.9 ± 5.0% in WT-VECs; p < 0.0001) and DNA damage (85.40 ± 7.42% in MAD-VECs vs 23.39 ± 6.11% in WT-VECs; p < 0.0001) (Fig. 1f). The NSCs generated from MAD-iPSCs were validated by sphere culture and expression of NESTIN, PAX6, and SOX2 (Supplementary Fig. 4a–c, j). No significant differences were observed either in the differentiation efficiency between MAD-iPSCs and WT-iPSCs or in the proliferative capability within 20 passages between MAD-NSCs and WT-NSCs (Fig. 1g and Supplementary Fig. 4c). Taken together, these data revealed defects in multiple lineages in this MAD patient as a result of the LMNA p.R527C mutation. The compromised vascular system was in line with the clinical features such as high blood pressure and atherosclerosis that are observed in MAD6,37.
MAD-MSCs recapitulated accelerated cellular senescence
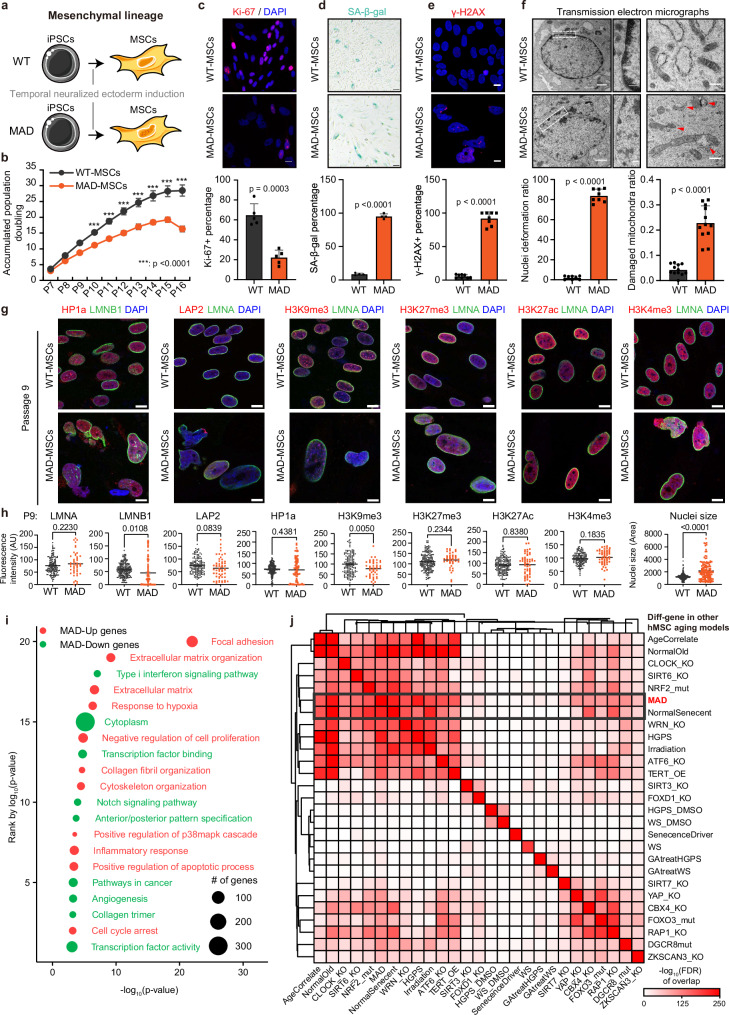
Stem cell decline38 is an essential contributor to many age-related morbidities39,40. The deterioration of vascular cells in MAD prompted us to examine the progenitor/stem cells, particularly the MSCs. The MSCs we generated satisfied the criteria of surface markers and tri-lineage differentiation capability (P5 MSCs) (Fig. 2a and Supplementary Fig. 5a, b). The MAD patient iPSCs-derived MSCs(hereafter referred to as MAD-MSCs) exhibited a shorter cellular lifespan, impaired proliferative capability, and accelerated senescence (P13 MSCs) (Fig. 2b–d), which were accompanied by the upregulation of cell cycle arrest genes p16 and INK4b as well as the activation of senescence-associated inflammasome (CXCL1, VEGFA, IL6, IL8, PAI, BMP2, PAI and BMP2) (P13 MSCs) (Supplementary Fig. 6c, d). As expected, MAD-MSCs manifested increased nuclear blebbing and DNA damage (P13 MSCs) (Fig. 2e and Supplementary Fig. 6b, e). Of note, MAD-MSCs exhibited enlarged nuclei (Fig. 2g, h and Supplementary Fig. 6f). Transmission electron microscopy (TEM) revealed a loss of peripheral heterochromatin in P13 MAD-MSCs (Fig. 2f). Furthermore, there was a significant increase in the percentage of mitochondrial damage in P13 MAD-MSCs (22.6% in MAD-MSCs vs 4.1% in WT-MSCs, p < 0.0001) (Fig. 2f). These data indicated MSCs from the MAD patient recapitulated accelerated cellular senescence.
Fig. 2. MAD-iPSCs derived mesenchymal stem cells (MAD-MSCs) recapitulated accelerated cellular senescence.
a, Human MSCs derived from WT and MAD-iPSCs by a temporal neuralized ectoderm induction method. b Growth curve of MSCs. Bars represent the mean ± SD.; n = 3 independent biological replicates; ***p < 0.001; n.s., non-significant; p value was calculated using two-way ANOVA test. c Proliferative capability measured by Ki 67 (red) using P13 MSCs. DAPI, blue. Scale bar 10 μm. Data represent the mean ± SD, n = 5. d SA-β-gal staining of MSCs at passage 13; Scale bar 100 μm. Data are mean ± SD, n = 3. e Representative image of γ-H2A.X (red) immunostaining at passage 13. DAPI, blue. Scale bar 10 μm. Data are mean ± SD, n = 8. The p values were calculated using two-tailed unpaired t-test. Experiments in c-e were repeated three times with similar results. f Representative transmission electron micrographs (TEM) of P13 WT- and MAD-MSCs. Scale bar 250 nm. The percentage of damaged mitochondria was quantified and calculated in MSCs. Data are mean ± SD, n = 462 WT, n = 368 MAD. The p value was calculated using two-tailed unpaired t-test. Three independent replicates were performed with similar results. g Representative images of co-staining of lamin B1 (LMNB1, green) and HP1a (red), lamin A/C (LMNA, green) and LAP2 (red), lamin A/C (LMNA, green) and H3K9me3 (red), lamin A/C (LMNA, green) and H3K27me3 (red), lamin A/C (LMNA, green) and H3K27ac (red), and lamin A/C (LMNA, green) and H3K4me3 (red) in passage 9 MSCs. DAPI, blue. Scale bars 10 µm. h Fluorescence intensity of g were quantified, including Lamin B1 and HP1a (WT n = 189, MAD n = 79), lamin A/C and LAP2 (WT n = 84, MAD n = 51), H3K9me3 (WT n = 88, MAD n = 64), H3K27me3 (WT n = 71, MAD n = 31), H3K27ac (WT n = 163, MAD n = 55), H3K4me3 (WT n = 145, MAD n = 79) and nuclei size (WT n = 293, MAD n = 143). Data are mean ± SD; the lines in scatter dot plot indicate the averaged intensity and the p values were calculated using two-tailed unpaired t-test. Three independent biological experiments were performed with similar results. i GO and KEGG enriched signaling pathways in MAD-MSCs. All p-values were determined by two-sided modified Fisher’s exact test using DAVID. j Comparison of aging-associated gene profilings between MAD-MSCs and other human MSCs aging models. Color depth indicates the level of transcriptional similarity. Source data are provided as a Source Data file.
To investigate early epigenetic changes behind MAD-MSCs aging, we examined the nuclear integrity and histone modifications at P9 when MAD-MSCs have not yet undergone senescence (Fig. 2b and Supplementary Fig. 6a). Significantly decreased lamin B1 and H3K9me3 were shown in MAD-MSCs compared with the control (Fig. 2g, h). Although the loss of LAP2a and HP1a were observed in multiple senescence conditions and MAD fibroblasts (Fig. 1d), significant changes in neither protein were observed in MAD-MSCs at P9 (Fig. 2g, h), but appeared at late passage P13 (Supplementary Fig. 6f). This suggests that loss of LAP2a and HP1a were not the initial response for senescence in MAD scenario. In addition, H3K9m3/H3K27me3-enriched heterochromatin budded off from the nuclei in MAD-MSCs (Supplementary Fig. 6e), which was in line with previous reports in different human MSCs senescence models41,42. Strikingly, the active histone markers H3K4me3 and H3K27ac were also found in regions of the budded nucleus and were even at times scattered in the cytoplasm (Supplementary Fig. 6e), reflecting the dramatically disorganized nucleus in MAD-MSCs. These findings suggested that accelerated stem cell senescence in this MAD patient could be a consequence of collapsed homeostasis in the nucleus.
Transcriptome comparison between MAD-MSCs and other human MSCs aging models
To investigate the potential contributors to MAD-MSCs senescence, RNA-seq was performed and compared between MAD-MSCs and WT-MSCs derived from the WT-iPSCs both at P9. Overall, 1488 genes were downregulated, while 1447 were upregulated in MAD-MSCs (two-fold change, p < 0.05) (Supplementary Fig. 7a, b; Supplementary Data 3). GO term analysis revealed the enrichment of extracellular matrix (ECM) organization (n = 39, p = 6.59E-10), negative regulation of cell proliferation (n = 49, p = 1.50E-05), cell cycle arrest (n = 21, p = 6.57E-04), cytoskeleton organization (n = 26, p = 3.29E-05) and inflammatory response (n = 43, p = 3.65E-04) in the upregulated genes, while notch signaling pathway (n = 19, p = 1.10E-04), transcription factor binding/activity (n = 80, p = 8.40E-04), angiogenesis and cancer-associated pathways were enriched in the downregulated genes (Fig. 2i). Interestingly, analysis of genetic assoicated diseases (GAD) database revealed that genes associated with cardiovascular disorders and metabolism disturbance were enriched in MAD-MSCs (Supplementary Fig. 7c, d).
To explore the similarities and differences between MAD-MSCs and other senescence models, we cross-analyzed 26 aging hMSCs transcriptomic datasets, including 20 premature senescent hMSCs and 6 senescence-delayed hMSCs models, termed ‘accelerated hMSCs’ and ‘alleviated hMSCs’, respectively (Supplementary Fig. 8a). Senescence, a process of cellular deterioration marked by chronic loss of homeostasis in macromolecules, is associated with increased expression of senescence-promoting genes and decreased expression of anti-senescence-associated genes. Based on this assumption, we defined the upregulated genes in senescent hMSCs and downregulated genes in alleviated hMSCs as senescence-associated genes, whereas the downregulated genes in senescent hMSCs and upregulated genes in alleviated hMSCs as geroprotection-associated genes (Supplementary Fig. 8b, c). The intersection of these gene sets revealed that different hMSCs aging models exhibited distinct transcriptional profiles and clustering of some datasets, indicating the aging process is complicated while interconnected (Fig. 2j). Notably, MAD-MSCs exhibited the highest transcriptional similarity to bone marrow-derived hMSCs undergoing replicative aging (Fig. 2j and Supplementary Fig. 8d).
LADs reorganization in MAD-MSCs
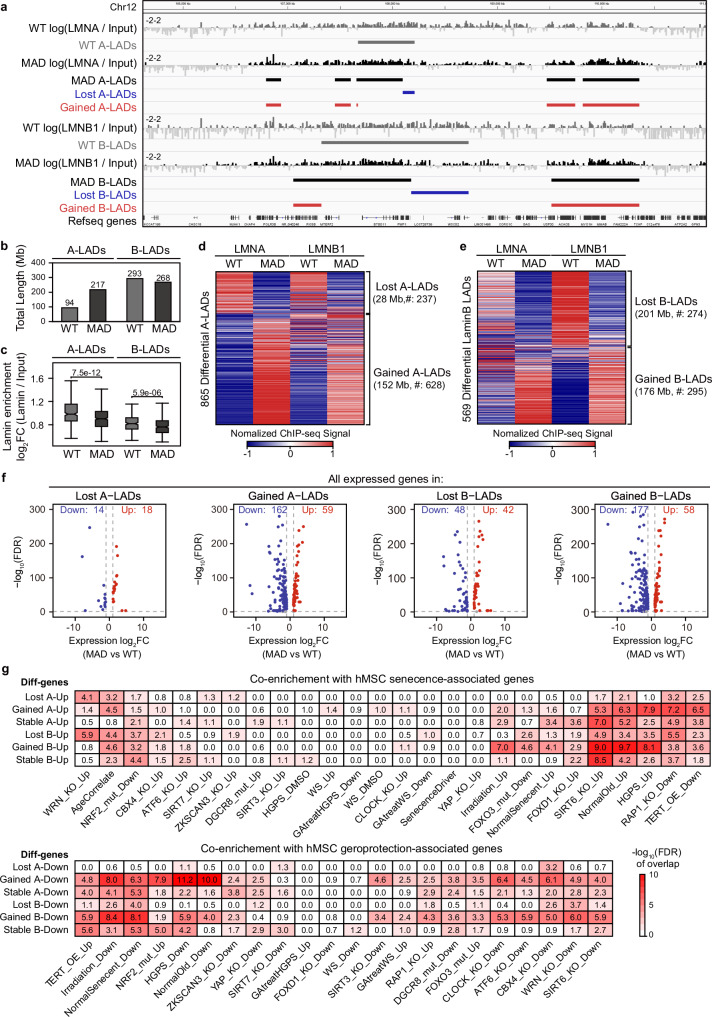
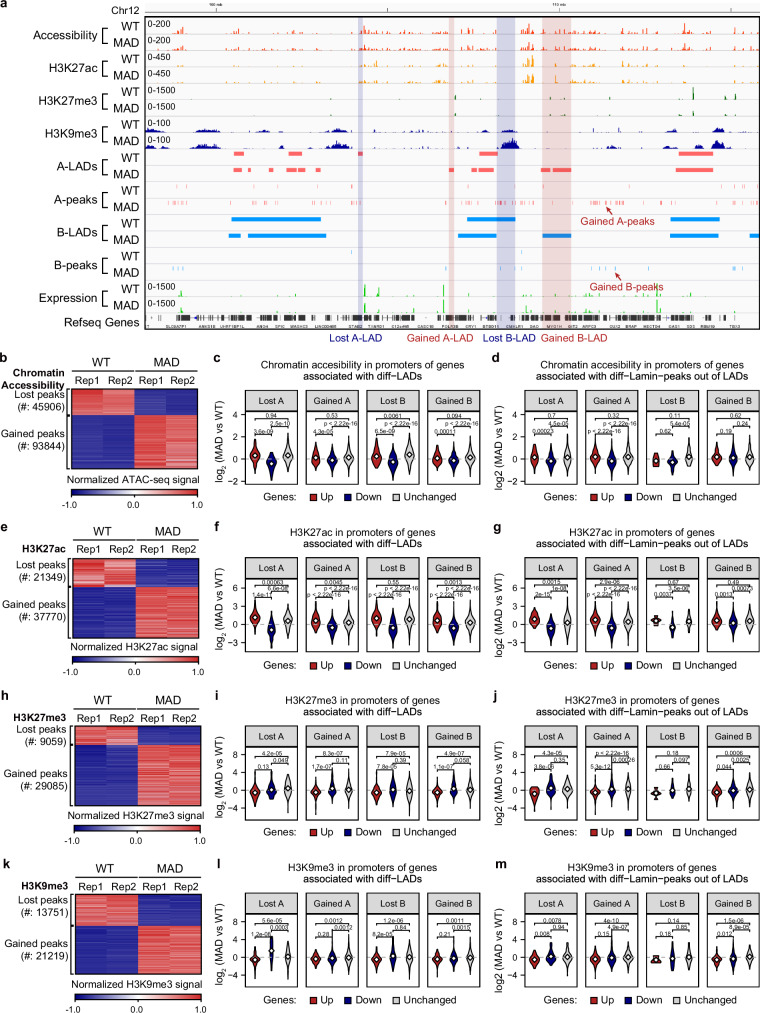
LADs reorganization has been reported in several laminopathy-based disorders, varying by cell-context20,23,43,44. It is well known that loss of lamin B1 is a biomarker of senescence, and lamin B1 LADs reorganization is involved in various aging models45,46. We performed both lamin A/C and lamin B1 chromatin immunoprecipitation and sequencing (ChIP-Seq) in WT and MAD-MSCs (MSCs at P9-P11). The lamin A/C LADs (A-LADs) and lamin B1 LADs (B-LADs) were identified by Enriched Domain Detector47 and further classified into three categories, e.g. loss, overlap, and gain, according to the specific change of genomic regions in MAD-MSCs when compared with WT-MSCs (Fig. 3a). Genome-wide comparison showed an increased number of both A- and B-LADs (237 lost vs 628 gained in A-LADs; 274 lost vs 295 gained in B-LADs) in MAD-MSCs (Fig. 3d, e). The genomic coverage of A-LADs increased dramatically from 94 Mb in WT-MSCs to 217 Mb in MAD-MSCs, whereas the B-LADs coverage was slightly decreased (293 Mb in WT-MSCs vs 268 Mb in MAD-MSCs) (Fig. 3b). Of note, the enrichment strength of both A- and B-LADs decreased significantly in MAD-MSCs (Fig. 3c). Overall, 865 differential A-LADs and 569 differential B-LADs were identified in MAD-MSCs compared with WT-MSCs (Fig. 3d, e).
Fig. 3. LADs reorganization is linked to aging-associated genes in MAD-MSCs.
a Representative distribution of LADs at specific genome loci in WT and MAD-MSCs, including A-LADs and B-LADs. b The length of genomic coverage of A-LADs and B-LADs in WT and MAD-MSCs. c Boxplot showing Lamin enrichment in WT A-LADs (n = 340), MAD A-LADs (n = 535), WT B-LADs (n = 257), and MAD B-LADs (n = 326), with 2 biological replicates for lamins ChIP-seq. Box plots display the median as the center line, the 25th and 75th percentiles as the bounds of the box, and the whiskers represent the minimum and maximum values within 1.5 times the interquartile range from the lower and upper quartiles. All p-values were determined using the two-sided Wilcoxon rank-sum test. d, e Heatmap of differential A-LADs and B-LADs in WT and MAD-MSCs. f Differentially expressed genes associated with LADs reorganization. Genes with over 2-fold transcriptional changes were counted. g Cross-analysis of the enrichment of dysregulated genes due to LAD reorganization in MAD-MSCs with geroprotection/senescence-associated profile in different hMSCs aging models. Color depth indicates enrichment score.
To further investigate the effects of LADs reorganization on transcriptional regulation, we integrated the transcriptome with differential LADs. Overall, 253 genes were dysregulated in reorganized A-LADs, while 318 were found in B-LADs (Fig. 3f; Supplementary Data 4). Notably, the gained LADs were associated with the majority of the transcriptomic changes when compared to the lost regions (221 genes in gained A-LADs vs 32 genes in lost A-LADs; 235 genes in gained B-LADs vs 90 genes in lost B-LADs) (Fig. 3f; Supplementary Data 4). To further evaluate the effect of LADs reorganization on MSCs aging, the dysregulated genes were cross-analyzed with senescence/geroprotection-associated gene profiles in hMSCs aging models. The expression changes observed in this MAD case, potentially mediated by LAD reorganization, were co-enriched with a variety of hMSCs models, especially normal old hMSCs and HGPS-MSCs (Fig. 3g). Notably, the gained LADs-associated genes were dominant for the co-enrichment of senescence- and geroprotection-associated profile (Fig. 3g).
For example, we observed a correlation between the downregulation of HDAC4, a gene associated with geroprotection48, and the gain of A-LADs in MAD-MSCs (2.6-fold decrease, FDR = 1.17e-38)(Supplementary Fig. 9a). CDK18, a cell cycle checkpoint factor safeguarding genome integrity49, was repressed in MAD-MSCs as it fell into the gained A-LADs (12.5-fold decrease, FDR = 3.20e-155) (Supplementary Fig. 9a). Similarly, several potential geroprotection-associated genes, including FOXC1 (22.5-fold decrease, FDR = 0), RNF130 (22.5-fold decrease, FDR = 0), ZNRF2 (6.2-fold decrease, FDR = 3.02E-68) and PRMT3 (22.5-fold decrease, FDR = 0), were downregulated as a result of gain of LADs (Supplementary Fig. 9a). Interestingly, lineage associated genes were also found to be dysregulated as LADs reorganization. Neural-related NEDD9 (4.1-fold increase, FDR = 1.50E-82), ANXA1 (2.6-fold increase, FDR = 1.13E-59), ANXA6 (2.1-fold increase, FDR = 1.13E-59), and immune-related IL7R (5.9-fold increase, FDR = 2.53E-121) and TRBC2 (5.1-fold increase, FDR = 6.32E-24), were found to be abnormally activated as a result of loss in LADs (Supplementary Fig. 9a). The data collectively suggest an association between extensive reorganization of LADs and altered expression profiles of genes that are involved in aging and lineage specification in MAD-MSCs.
Reposition of non-LAD lamina-chromatin binding peaks participated in aging-associated gene regulation in MAD-MSCs
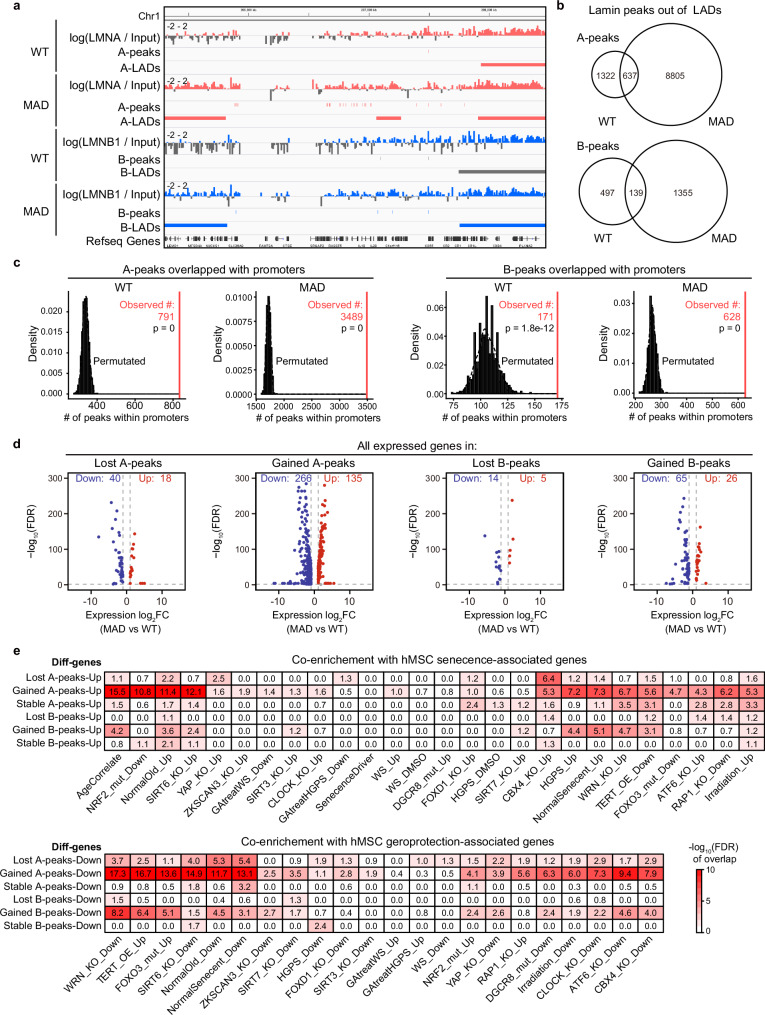
When interpreting lamina-chromatin interactions, we identified many non-LAD lamina-chromatin binding peaks, termed ‘peaks out of LAD’, that were dispersed outside of LADs (Fig. 4a). Strikingly, there were a total of 1959 peaks out of A-LADs in WT-MSCs and 9442 peaks out of A-LAD in MAD-MSCs, with only a small portion (637 peaks) overlapping (Fig. 4b). Likewise, 636 and 1494 peaks out of B-LADs were identified in WT-MSCs and MAD-MSCs respectively, with only 139 shared peaks (Fig. 4b). To further investigate the potential function of these non-LAD lamina-chromatin interaction peaks, genome-wide co-occurrence between non-LADs lamina-chromatin binding peaks and promoters were analyzed. Notably, non-LADs peaks were not randomly repositioned but rather co-occurred with the promoter (permutation test, p = 0) (Fig. 4c), suggesting that non-LAD peaks were likely involved in transcriptional regulation. Indeed, a total of 569 differential genes fell into the repositioned non-LADs peaks in MAD-MSCs (Fig. 4d; Supplementary Data 4). Among these genes, the gained peaks out of A-LADs accounted for the majority of the changes (401 genes, 77.1%), while the gained peaks out of B-LADs contributed 16.0% (91 genes) to the changes (Fig. 4d). These dysregulated genes were further cross-analyzed based on senescent and geroprotective gene profiles to evaluate the co-enrichment between repositioned non-LADs peaks and senescence. Notably, the gained peaks out of A-LADs exhibited the highest enrichment in upregulated senescence-associated genes and downregulated geroprotection-associated genes across the different hMSCs aging models (Fig. 4e).
Fig. 4. Reposition of non-LAD lamina-chromatin binding peaks modulates aging-associated genes in MAD- MSCs.
a Distribution of non-LADs lamina-chromatin binding peaks at specific genome loci in WT and MAD-MSCs. Each vertical bar represents one peak out of LADs. b Global non-LAD lamina-chromatin binding peaks identified in WT and MAD-MSCs. The number of the peaks are indicated. c Genome-wide co-occurrence of non-LAD lamina-chromatin binding peaks with promoters using a one-sided permutation test. The vertical axis density represents the frequency of co-occurrence of non-LAD binding peaks with promoters while horizonal axis represents predicted co-occurrence number. The observed co-occurrence number are indicated. d Differentially expressed genes associated with repositioned non-LADs lamina-chromatin binding peaks in MAD-MSCs Genes with over 2-fold transcriptional changes were counted. e Cross-analysis of enrichment of dysregulated genes due to reposition of non-LAD lamina-chromatin binding peaks in MAD-MSCs with geroprotection/senescence-associated profile in different hMSCs aging models. Color depth indicates the enrichment score.
TBX2, an indispensable transcription factor for early development and geroprotection50, showed decreased expression in MAD-MSCs, corresponding to the increased lamina-chromatin binding peaks in MAD-MSCs (5.1-fold decrease, FDR = 1.09E-226) (Supplementary Fig. 10a). DCP2, a protein responsible for mRNA decapping, RNA decay and closely associated with the aging process51,52, was downregulated in MAD-MSCs, possibly due to the gained lamina-chromatin binding peak in its promoter (2.1-fold expression decreases, FDR = 9.17E-27) (Supplementary Fig. 10a). Similarly, CEP70, a centrosomal protein critical for microtubules organization during mitosis, was downregulated in MAD-MSCs, which could be attributable to the gained LAD and lamina-chromatin binding peak at its promoter (2.7-fold decrease, FDR = 1.57E-33) (Supplementary Fig. 10a). Interestingly, several lineage specific genes, including NNAT (5.2-fold increase, FDR = 3.45E-60) and NAV2 (2.8-fold increase, FDR = 1.01E-82), were also abnormally activated in repositioned lamina-chromatin binding peaks (Supplementary Fig. 10a). These observations suggest that in addition to LADs reorganization, the repositioning of non-LAD lamina-chromatin binding peaks may also be associated with changes in transcriptional regulation, potentially influencing MAD-MSCs senescence and aspects of progeroid pathogenesis.
Lamina-chromatin interaction coordinated with chromatin features to regulate transcription in MAD-MSCs
To explain transcriptional upregulation in the gained lamina-chromatin interaction sites and transcriptional downregulation in the lost lamina-chromatin binding regions (Figs. 3e and 4e), we integrated multiple profiling data, including active marks A transposase-accessible chromatin with sequencing (ATAC-Seq) and H3K27ac ChIP-Seq as well as repressive marks H3K9me3 and H3K27me3 ChIP-Seq, to examine the contribution of chromatin features to the transcriptional dysregulation in MAD-MSCs compared to WT-MSCs (Fig. 5a). ATAC-Seq revealed a more open chromatin state in MAD-MSCs with 45,906 lost peaks and 93,844 gained peaks (FDR < 0.05) (Fig. 5b). Consistently, H3K27ac ChIP-Seq identified 21,349 lost peaks and 37,770 gained peaks in MAD-MSCs, corresponding to 31,478 inactivated typical enhancers (TEs), 188,270 activated TEs, 560 inactivated and 264 activated super-enhancers (SEs) (Fig. 5e and Supplementary Fig. 11c-g). Globally, the ATAC signals were mainly located in intron (48%) and intergenic regions (41%). About 20% of ATAC was associated with the enhancer, and 11% occurred at LADs (Supplementary Fig. 11a, 11h). Of note, 22% of the lost ATAC peaks in MAD-MSCs were enriched at promoter regions and more than half were mapped to the enhancer regions, especially SEs (46%) (Supplementary Fig. 11h). Meanwhile, 11% of gained ATAC peaks in MAD-MSCs were associated with enhancers (Supplementary Fig. 11h). In lost ATAC peaks, 6% were mapped to A-LADs and 9% was mapped to B-LADs. In the gained ATAC peaks, 4% were mapped to A-LADs and 12% to B-LADs (Supplementary Fig. 11a). About 9% of gained ATAC fell in lamin B1-specific LADs, in contrast to 1% in LAMIN A-specific LADs (Supplementary Fig. 11a). The altered enhancers in MAD-MSCs exhibited a pattern similar to that of ATAC in the distribution of lamin A/C- and lamin B1-LADs (Supplementary Fig. 11a, b). Interestingly, the enriched ATAC motifs in MAD-MSCs were highly matched with the pioneer transcriptional factors binding regions enriched in HGPS fibroblasts53 (Supplementary Fig. 11i). When integrated with transcriptomic profiling, it was noted that ATAC and H3K27ac signals were significantly higher in the promoter regions of upregulated genes compared with the promoter regions of downregulated genes, regardless of the lamina-chromatin interaction states (Figs. 5c, d, 5f, g).
Fig. 5. Lamina-chromatin interaction coordinates with chromatin features to regulate gene expression.
a Representative distribution of different chromatin features in lamina-chromatin binding sites. b Overall view of ATAC peaks redistribution in MAD-MSCs. c Averaged chromatin accessibility in promoter regions with different A and B-LADs reorganization. d Averaged chromatin accessibility in the promoter region with different non-LAD lamina-chromatin binding peaks. e Overall view of H3K27ac peaks redistribution in MAD-MSCs. f Averaged H3K27ac peaks in promoter regions with different A and B-LADs reorganization. All p-values were determined using the two-sided Wilcoxon rank-sum test. g Averaged H3K27ac peaks in promoter regions with different non-LAD lamina-chromatin binding peaks. All p-values were determined using the two-sided Wilcoxon rank-sum test. h Overall view of H3K27me3 redistribution in MAD-MSCs. i Averaged H3K27me3 peaks in the promoter regions with different A and B-LADs reorganization. All p-values were determined using the two-sided Wilcoxon rank-sum test. j Averaged H3K27me3 peaks in promoter regions with different non-LAD lamina-chromatin binding peaks. All p-values were determined using the two-sided Wilcoxon rank-sum test. k Overall view of H3K9me3 redistribution in MAD-MSCs. l Averaged H3K9me3 peaks in promoter regions with different A and B-LADs reorganization. All p-values were determined using the two-sided Wilcoxon rank-sum test. m Averaged H3K9me3 peaks in promoter regions with different non-LAD lamina-chromatin binding peaks. All p-values were determined using the two-sided Wilcoxon rank-sum test.
In parallel, H3K27me3 peaks and H3K9me3 signals were also found to be increased (13,751 lost vs 21,219 gained and 9059 lost vs 29,085 gained, respectively) in MAD-MSCs (Fig. 5h, k), with a distribution pattern that differs to ATAC and H3K27ac peaks (Supplementary Fig. 11j–k). Interestingly, H3K27me3 and H3K9me3 signals were significantly higher in the promoter regions of down-regulated genes compared with the promoter regions of up-regulated genes, which is independent of the lamina-chromatin interaction states (Figs. 5i–j, 5l–m).
The lower signals of ATAC and H3K27ac and higher H3K27me3 signals in HDAC4 and CDK18 gene loci may explain the downregulation of geroprotection-associated HDAC4 and CDK18 within gained A-LADs in MAD-MSCs (Supplementary Fig. 12a). Similarly, down-regulation of ZSCAN12 (2.8-fold decrease, FDR = 4.74E-16) and DHRS3 (42.3-fold decrease, FDR = 1.93E-292) were observed in gained B-LADs with lower signals of ATAC and H3K27ac and higher signals of H3K9me3 and H3K27me3 (Supplementary Fig. 12b). On the other hand, loss of A-LADs occupancy in NEDD9 locus with higher ATAC and H3K27ac signals at the promoter region may have contributed to the activation of a lineage specific gene (Supplementary Fig. 12a). Lower signal of repressive marks and higher signal of active marks enriched in the lost LADs resulted in up-regulation of NTM (27.1-fold increase, FDR = 0) and NEK7 (5.2-fold increase, FDR = 4.93E-266) (Supplementary Fig. 12c). The PDGFA locus, located in the gained LAD in MAD-MSCs with a 4.9-fold increase in transcription (FDR = 4.53E-93), exhibited a higher signal of ATAC and H3K27ac but lower signal of H3K9me3 (Supplementary Fig. 12d). In the FBXO4 locus, the lower active marks explained why FBXO4 expression decreased in MAD-MSCs (2.5-fold decrease, FDR = 9.38E-28) even though LAD occupancy was lost (Supplementary Fig. 12d). The regulation of transcription by integrated chromatin features were also observed in genes located around non-LADs lamina-chromatin binding peaks, such as KAT2B, MYO6, SMAD2 and HIPK2 (Supplementary Fig. 13). Taken together, these observations suggest the transcription landscape observed in MAD-MSCs could be a coordinated action of lamina-chromatin interaction state and specific local chromatin features.
Hierarchical chromatin disorganization potentially shapes stem cell aging
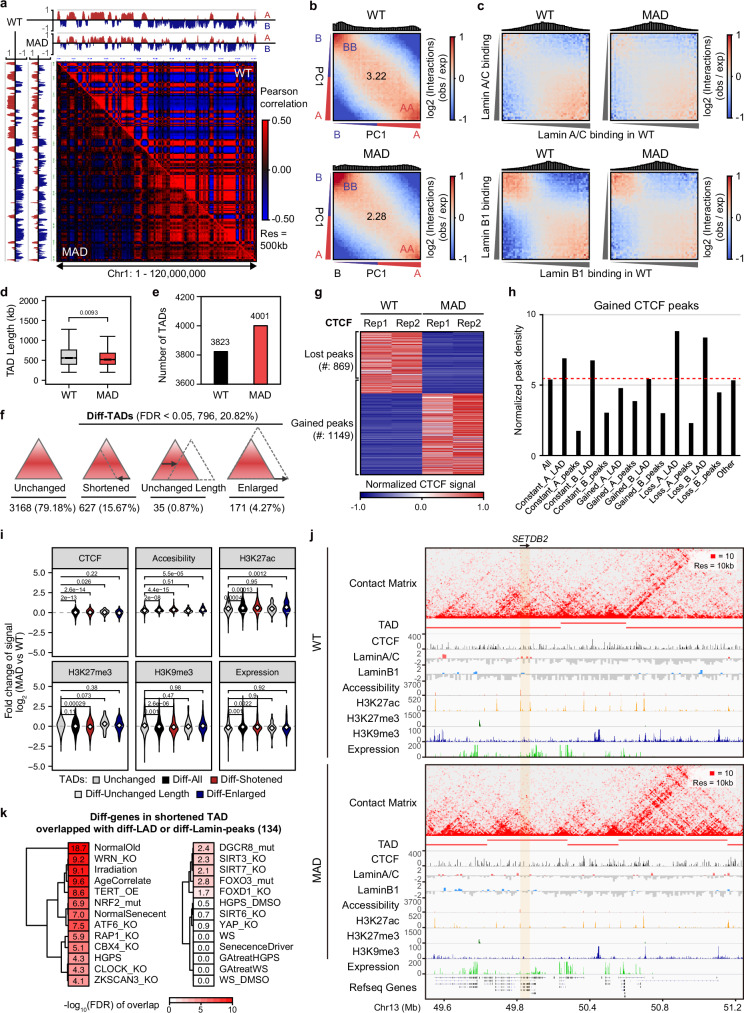
Given that nuclear lamins facilitate the establishment of high-order chromatin architecture19,24, we then interrogated the chromatin conformation change in MAD-MSCs. Hi-C was performed with two biological replicates for each sample (Supplementary Fig. 14a), the statistics of the relevant datasets were summarized in Supplementary Data 7, and high-resolution of chromosome conformation was obtained (Supplementary Fig. 14c). In MAD-MSCs, an increase in short-distance interaction frequency and a decline in long-distance interaction frequency were observed (Supplementary Fig. 14b). While clear segregation of compartment A and B was observed in WT-MSCs, the separation became largely indistinct in MAD-MSCs (Fig. 6a and Supplementary Fig. 15). Saddle plot analysis indicated a global loss of compartmentalization, quantifying the strength of chromatin compartment based on the interaction frequencies arranged by eigenvector. The interactions of compartment A-B and B-B were increased, while the interactions of compartment A-A were decreased in MAD-MSCs (WT_AA:1.266, MAD_AA:1.109; WT_BB:1.270, MAD_BB1.308; WT_AB:0.707, MAD_AB:0.796) (Fig. 6b). When lamins-associated chromatin was analyzed, the interactions of the lamin-binding chromatins were significantly decreased in MAD-MSCs (Fig. 6c), suggesting that MAD mutation in LMNA possibly impairs high order chromatin organization and compartmentalization through jeopardized the chromatin interaction. Additionally, we observed 4.66% of compartment A converted to B and 14.36% of compartment B switched to A, in addition to the compartmental strengthening and weakening (Supplementary Fig. 14d). The higher percentage of compartment B to A switching was consistent with chromatin features observed in MAD-MSCs, including increased ATAC and H3K27ac marks.
Fig. 6. MAD mutation associates with loss of chromatin compartmentalization and increase in TADs.
a Normalized heatmap of specific region (q arm of chromosome 1) in WT and MAD-MSCs. The color maps of relative interaction probability in WT-MSCs and MAD-MSCs were displayed on the same scale. The A and B compartments were defined by PC1 signal (positive PC1 regions in red color represent A compartments, negative PC1 regions in blue color represent B compartments). b Statistical analysis of compartment interaction between compartment A and compartment B in WT-MSCs and MAD-MSCs according to Saddle plot analysis. c Analysis of the lamina-compartment interactions. d Boxplot showing length of WT TADs (n = 3823) and MAD TADs (n = 4001), with 2 biological replicates for Hi-C. Box plots display the median as the center line, the 25th and 75th percentiles as the bounds of the box, and the whiskers represent the minimum and maximum values within 1.5 times the interquartile range from the lower and upper quartiles. All p-values were determined using the two-sided Wilcoxon rank-sum test. e Overall view of TAD number in WT-MSCs and MAD-MSCs. f Category of differential TADs number presented in MAD-MSCs, including stable, shortened, shifted without change in length, and enlarged. g Overall view of CTCF redistribution in MAD-MSCs. h The distribution of CTCF across lamin-chromatin interaction sites. i Overall view of altered chromatin features, including CTCF binding, ATAC, H3K27ac, H3K27me3, H3K9me3, and gene expression in TADs. All p-values were determined using the two-sided Wilcoxon rank-sum test. j Integrative analysis of TAD disorganization and chromatin features in genomic region covering a dysregulated aging-associated gene, SETDB2, in MAD-MSCs. k Cross-analysis of the enrichment of dysregulated genes resulted from shortened TADs in MAD-MSCs with geroprotection/senescence-associated profile in hMSCs aging models. Color depth indicates enrichment score.
TopDom54 analysis revealed 3823 and 4001 TADs in WT-MSCs and MAD-MSCs, respectively (Fig. 6e). The increased TAD number in MAD-MSCs was accompanied by a shorter TAD length (725 kb on average in WT-MSCs vs 683 kb on average in MAD-MSCs) (Fig. 6d). The differential TADs were subsequently classified into three categories, with a significant portion of TADs (627) being either shortened (15.67%), shifted without length change (0.87%) or enlarged (4.27%), despite of 79.18% of stable structured TADs (FDR < 0.05) (Fig. 6f). CCCTC-binding factor (CTCF) is a core factor for TAD boundary maintenance and CTCF ChIP-Seq revealed the increased CTCF binding peaks in MAD-MSCs (Fig. 6g). The increased CTCF tended to bind at the A-LADs and B-LADs lost regions in MAD-MSCs which aligned with the boundaries of gained TADs (p = 0.0048) (Supplementary Fig. 16a), indicating that the mutant lamin A/C may contribute to TADs separation in MAD-MSCs. To further investigate the relationship between TADs reorganization and senescence, chromatin features and transcriptomic profiles were integrated with TAD structure. Notably, remarkable changes in ATAC, H3K27ac, CTCF binding, H3K9me3, as well as transcription were all observed in the shortened TADs (Fig. 6i). Higher gene expression within the shortened TADs has been noted previously55 and may be linked to their relatively higher level of accessibility. Further analysis identified 110 genes (Supplementary Data 8) located in regions overlapped by shortened TADs and reorganized LADs or altered non-LAD lamina-chromatin binding peaks, with the highest co-enrichment to senescence/geroprotection-associated profile in normal hMSCs aging (Fig. 6k). Among them, 80 were downregulated while 30 were upregulated (FDR < 0.05) (Supplementary Fig. 16b). For example, shortened TADs of SETDB2, a H3K9me3 methyltransferase, was accompanied by lower ATAC, reduced H3K27ac and decreased transcription in MAD-MSCs (3.4-fold decrease, FDR = 5.80E-66) (Fig. 6j). Similarly, PPARα, a member of the peroxisome proliferator-activated receptor regulating energy metabolism and oxidative stress during aging process, was repressed (3.2-fold decrease, FDR = 1.18E-85) (Supplementary Fig. 16d). In addition, KRT19, a type I cytokeratin mainly expressed in epidermis and involved in mesenchymal-epithelial transition (MET), was upregulated in MAD-MSCs (6.8-fold increase, FDR = 9.15E-281), possibly linked to the changes in shortened TADs and local chromatin state(Supplementary Fig. 16d). GO term analyzes revealed the enrichment in ECM regulation, collagen biogenesis, ossification, and DNA binding transcription factor activity in MAD-MSCs (Supplementary Fig. 16c). Dysregulation of ECM remodeling and collagen biogenesis are known to be involved in aging both in vitro and in vivo56,57. Acroosteolysis is an important feature of MAD while chronic bone loss is a typical clinical manifestation in normal aging. The enrichment of negative regulation of ossification associated with TAD shortening (Supplementary Fig. 16c) was cosistent with observed acroosteolysis in MAD.
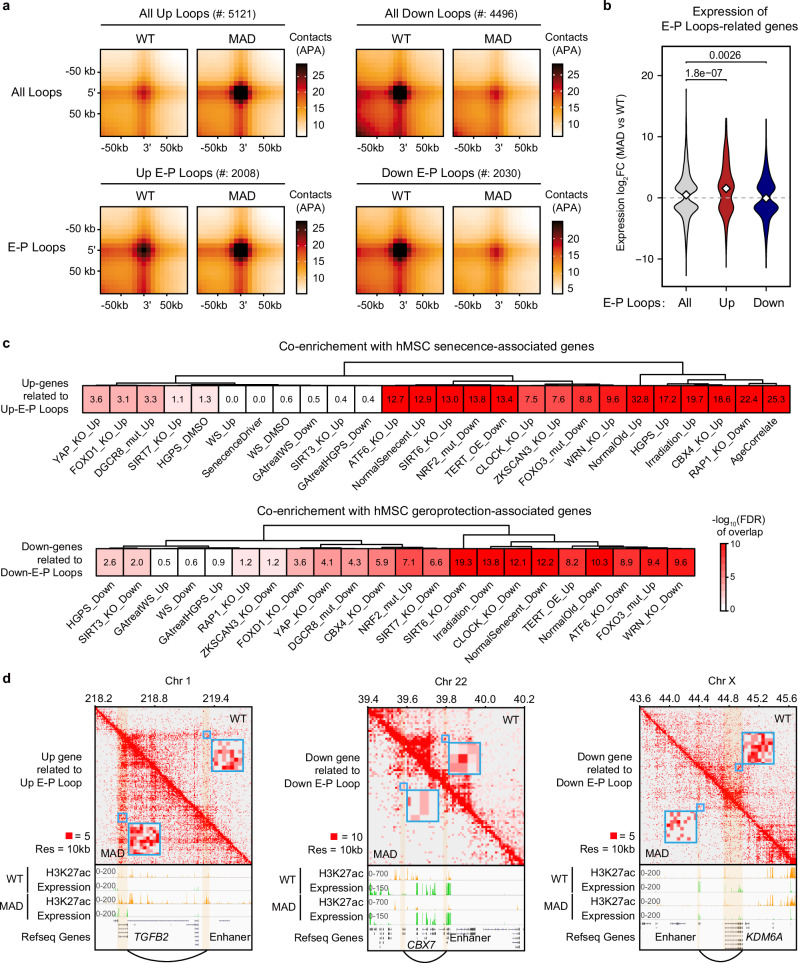
At the chromatin loop level, 5121 strengthened and 4496 weakened chromatin loops genome-wide were annotated in MAD-MSCs (Fig. 7a). Among which, 2008 and 2030 enhancer-promoter (E–P) loops were further identified based on H3K27ac signals (Fig. 7a). These differential E-P loops presented significant correlation with transcription regulation as strengthened E-P loops occupied regions of elevated gene expression (p = 1.8e − 07) while weakened E-P loops hold decreased transcription (p = 0.0026) (Fig. 7b), corresponding to 126 upregulated genes and 99 downregulated genes, respectively (Supplementary Fig. 17a; Supplementary Data 9). Intersection analyzes of the datasets revealed the highest co-enrichment in senescence-associated genes and geroprotection-associated genes between MAD-MSCs and hMSCs of normal aging (Fig. 7c). For examples, E-P loop was strengthened in TGFB2 locus with abnormal activation of TGF-β2 (29.7-fold increase, FDR = 0, n = 5) (Fig. 7d), a member of TGF-β family exhibiting inhibitory effect on cell proliferation and induces cellular senescence58. Conversely, CBX7, a geroprotection associated gene encoding a component of the polycomb group PRC1-like complex59, was decreased (4.4-fold decrease, FDR = 1.12E-84, n = 10) due to weakened E-P loop (Fig. 7d). Interestingly, E-P loop of KDM6A locus was weakened in MAD-MSCs with a decreased expression of KDM6A (2.1-fold decrease, FDR = 5.71E-40, n = 5), a demethylase of H3K27me3 (Fig. 7d). The reduced KDM6A in MAD-MSCs may explain why global loss of H3K27me3 was not observed in MAD-MSCs, contrary to the reduced H3K9me3. In addition, dysregulation of senescence-associated gene, such as MMP1460 (3.2-fold increase, FDR = 1.66E-161, n = 5), (Supplementary Fig. 17d), fell into the altered E-P loop. Additionally, several other potential aging-associated genes resulted from E-P loop alterations were also annotated, including TBX2 (5.1-fold decrease, FDR = 1.09E-226, n = 14), PCK2 (2.8-fold decrease, FDR = 1.18E-41, n = 5) and MMP16 (59.5-fold increase, FDR = 0, n = 5) (Supplementary Fig. 17d). These data indicate that mutant lamin A/C is associated with global disorganization of the chromatin hierarchy, potentially contributing to accelerated aging in stem cells by influencing the regulation of genes involved in epigenetic modification, senescence, and geroprotection.
Fig. 7. Altered chromatin E-P loops in MAD-MSCs are implicated in stem cell aging.
a Statistical analysis of global changes in chromatin loop and enhancer-promoter loop. b The transcription changes of altered E-P loops in MAD-MSCs. All p-values were determined using the two-sided Wilcoxon rank-sum test. c Cross-analysis of the enrichment of dysregulated genes resulted from E-P loop alteration in MAD-MSCs with geroprotection/senescence-associated profile plotted by hMSCs aging models. Color depth indicates enrichment score. d Representative transcriptional dysregulation corresponds with strengthened and weakened E-P loops, respectively.
Discussion
APSs caused by LMNA mutations are underrepresented in the literature, compared with the intensively studied TPS. In this study, iPSCs were established from a male patient with MAD, a representative APS with a specific LMNA mutation (c.1579 C > T, p. R527C), to investigate the changes in chromatin organization and the mechanism underlying the pathogenesis of MAD. In addition to LADs, dramatic changes leading to dysregulation of senescence- and geroprotection-associated genes were also observed in non-LADs lamina-chromatin binding peaks, whose alteration has not received much attention. Examination of the chromatin conformation revealed a hierarchical disorganization is an intrinsic force modulating stem cell aging in MAD.
Consistent with previous reports7,61, senescence-associated defects observed in parental MAD-fibroblasts were rejuvenated in iPSCs, implying that temporal activation of reprogramming factors may be a potentially effective strategy in ameliorating aging-related deterioration62. MAD mesenchymal lineages exhibited defects corresponding to cardiovascular dysfunction in progeria patients. LMNA mutation in MAD affects both lamin A and lamin C whereas the HGPS mutation only affects lamin A. It has been reported that neural-specific miR-9 can repress the expression of lamin A and progerin but not lamin C, providing an explanation for the absence of neurological defects in HGPS63,64. Given that neither neural cells defects nor neurological symptoms were observed in MAD patients, it is plausible that lamin C variants alone is insufficient to deteriorate neuronal cell functions.
Genome-wide LADs identification provides a comprehensive landscape of lamina-chromatin interaction65–67 and reveals close correlations between transcriptional regulation and lamina-chromatin contacts during development and disease pathogenesis20–22,43,68,69. In this study, we mapped both A and B-LADs in MAD-MSCs and found that the pathogenic mutation gave rise to a decreased strength in the lamina-chromatin interaction but increased LAD number and coverage. The gained LADs accounted for the majority of transcription changes commonly enriched in the senescence- and geroprotection-associated gene profiles, suggesting a critical role of gained LADs in driving MAD-MSCs aging (Fig. 3f, g). In addition, despite more than half of the genes were modulated by overlapped A-and B-LADs, considerable gene expression in MAD-MSCs were regulated independently by either A- or B-LADs (Supplementary Fig. 9b, c and Supplementary Data 6).
The fundamental changes in the lamina-chromatin interaction in MAD-MSCs also extended beyond LADs. The altered non-LADs lamina-chromatin binding peaks were not randomly distributed but rather preferentially occurred in the promoter regions (Fig. 4c). Specifically, the gained A-type non-LAD lamina-chromatin binding peaks that co-occurred with 3849 promoters were responsible for the dysregulation of nearly 400 genes in MAD-MSCs, representing the initial changes of MAD. In line with this observation, the altered pS22-lamin A/C binding peaks have been linked to the premature senescence in HGPS by co-enrichment of enhancers and c-JUN70. Therefore, transcription regulation by non-LAD lamina-chromatin binding peaks plays a critical role in MAD-MSCs aging and must be taken into consideration when dissecting the mechanisms behind the various laminopathies.
Interestingly, the gained lamina-chromatin interaction was not always accompanied by the decreased gene expression in MAD-MSCs (Figs. 3f and 4d). Conversely, loss of lamina-chromatin interaction did not necessarily lead to transcriptional activation (Figs. 3f and 4d), though to a lesser extent. Our data revealed that silencing or activation of transcription requires a coordinated action between lamina-chromatin interaction and active/repressive chromatin marks (Fig. 5 and Supplementary Fig. 12, 13). This is in line with previous observations that promoters within LADs can escape the intrinsic repression and the genes are not upregulated upon LADs loss65,67,68,71,72. These observations undermined a commonly held belief that lamina-chromatin binding is transcriptionally repressive. Our study highlights the importance of LADs and non-LADs lamina-chromatin binding peaks in gene regulation and disease pathogenesis.
Laminopathies are generally accompanied by loss of nuclear integrity closely linked to 3D genome disorganization. These hierarchical changes in MAD-MSCs represented the most drastic chromatin reorganization, compared to other senescence models23,32–35 reported so far where only subtle changes in chromatin compartmentalization and TADs were observed. We propose that the genome-wide compartmentalization loss, also observed in late passage of HGPS fibroblast, is associated with a decreased nuclear lamina-chromatin binding strength (Fig. 6a–c), supporting the concept that lamins are essential for the establishment of chromatin architecture24. Interestingly, Zheng et al. reported that the overall chromatin architecture and TADs profiling were preserved in triple lamin-depleted mESCs while only certain intra- and inter-TAD interactions were altered67. One plausible explanation of this discrepancy is that unidentified factors compensate lamins’ function in chromatin organization in mESCs but are degraded upon senescence as the activation of nuclear autophagy42,73,74.
Many hallmarks have been proposed, including cellular senescence, genome instability, epigenetic modification, intercellular communication alteration and mitochondria dysfunction38, and characterized as biomarker of aging, however, the driving force underlying all these hallmarks but not alone behind a specific aging model remains unclear. In this study, we found alterations of lamina-chromatin interaction were responsible for cell cycle related CDK18 and chromatin remodeler HDAC4 (Supplementary Fig. 12a). The cross-analysis of the shortened TADs and lamina-chromatin interaction identified 110 potentially dysregulated genes, including SETDB2 and PPARα (Fig. 7j). As the reduced H3K9me3, a downstream target of SETDB2, is characterized as a driving force for aging30 and was also observed in MAD-MSCs even at early passages (Fig. 2g), the decreased SETDB2 resulted from a shortened TAD therefore provides a new mechanistic explanation for the loss of heterochromatin and accelerated senescence in MAD-MSCs. The identification of reduced PPARα as reorganized TAD structure likely explains the mitochondria damages in MAD-MSCs (Fig. 2f and Supplementary Fig. 16d). In addition, alteration of chromatin loops revealed the dysregulation of several aging-associated genes (i.e., epigenetic factor CBX7) and development-associated genes, including TBX2, PDGFC and MYL12A (Fig. 7d and Supplementary Fig. 17d) from another layer. Notably, the dysregulation of epigenetics associated genes are identified in all levels of disorganized chromatin hierarchy. Taken together, our integrated omics analyzes systematically dissect the mechanisms underlying many different aging hallmarks in MAD-MSCs from the conception of chromatin hierarchy change.
More importantly, our study also revealed abnormal activation of multiple lineage specification genes in MAD-MSCs (Supplementary Figs. 9a, 10a, 16d, 17d). Abnormal expression of neural associated PAX6 and SOX2 has been reported in laminopathy-based cardiomyocytes43 and a very recent study reported that the reactivation of placenta-specific PSG4 in late passage of MSCs derives accelerated senescence75. Although it remains uncertain whether the sporadic expression of lineage-specific genes in MAD-MSCs influences senescence, it clearly indicates a disordered state of transcription influenced by altered chromatin architecture. Additionally, the activation of other lineage-specific genes in MSCs suggests a reason for the reduced differentiation potency observed in senescent MSCs, posing challenges to MSC-based therapies76–79.
Our study suggests that a pathogenic LMNA mutation may lead to changes in the organization of chromatin at multi-levels. These changes could affect gene activity and contribute to the aging of stem cells in laminopathy-based atypical progeria. While significant progress has been made in understanding the molecular alterations associated with the MAD mutation, it remains challenging to definitively distinguish the direct primary effects from the indirect secondary consequences.
This work systematically investigates the multidimensional chromatin conformation alterations underlying accelerated stem cell aging from atypical progeria MAD. Although we demonstrated the disorganized chromatin hierarchy plays a critical role in stem cells aging, manipulation of specific chromatin structure, such as fusing the separated TADs, has not been achieved to ameliorate aging. In addition, our study identified a number of genes potentially involved in ageing processes, such as PCK2, TBX2. Further characterization of these genes in aging both in vitro and in vivo would provide additional evidence supporting our findings and strengthen our conclusion.
Methods
Primary cell isolation and culture
Skin tissues were collected from a 3-year-old male MAD patient, 5-year-old male HGPS patient and a 26-year-old female healthy person with the informed consent given by guardians and under the guidance of ethical regulations in Dongguan Eighth People’s Hospital, Dongguan, China. Skin tissues were sterilized and cut into small pieces before transferred T25 flasks. Five milliliters of fibroblast culture media containing DMEM/HG, 10% human umbilical cord blood serum, 1 mM GlutaMax, and 1% Penicillin/ Streptomycin, was then added for culture and the expansion of spindle-like fibroblast cells was monitored daily. Human iPSCs were reprogrammed from fibroblasts using minicircle DNA vector as previously described36. Cells were maintained in a humid incubator at 37 °C and 5% CO2 and tested negative for mycoplasma infection based on PCR method by the Center for PanorOmic Sciences, Li Ka Shing Faculty of Medicine. We have obtained informed consent from all participants for the publication of information, including clinical information, that may identify them.
Teratoma formation
Undifferentiated iPSCs (about 3 × 106) were harvested and resuspended in a 50% Matrigel solution, and then injected into the abdominal cavity of female NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) mice, which were aged between 6–8 weeks. Teratomas were isolated after 8–12 weeks, fixed in 4% PFA, dehydrated in 70% ethanol, and embedded in paraffin. Sections were stained with hematoxylin and eosin (H&E) for analysis of mesoderm, ectoderm, and endoderm differentiation. The mice were housed in a 12 h light/12 h dark cycle at ~23 °C and 40% relative humidity. All mice experimental protocols were approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR) of The University of Hong Kong.
Generation of NSCs, VSMCs and VECs from human iPSCs
NSCs were derived from human iPSCs as discribed61. Briefly, iPSCs were dissected to single cells and plated in a Matrigel-coated 6-well plate overnight. The culture medium was switched to neural induction medium (50% DMEM/F12, 50% Neurobasal, 1% N2 supplement, 2% B27 supplement and 10 ng/ml of human LIF, 4 μM CHIR99021, 3 μM SB431542) for one week. NSCs were then split at 1:4 with NSCs maintenance medium (50% DMEM/F12, 50% Neurobasal, 1% N2 supplement, 2% B27 supplement, 2 mM GlutaMAX and 10 ng/ml of human LIF, 3 μM CHIR99021, 1 μM SB431542). NSCs were further characterized by NESTIN, PAX6 and SOX2 after additional 5 passages and then subjected to downstream assays. The VSMCs and VECs were generated from iPSCs as previously described80. Cells were firstly induced to mesoderm and then switched to VSMC induction medium (N2B27 basal medium plus 20 ng/ml PDGF-BB, 2 ng/ml Actvin A and 2 μM Heparin) or VECs induction medium (N2B27 basal medium plus 200 ng/ml VEGF-A and 2 μM Forskolin) for another 7–10 days. Generated VSMCs were further cultured in LG-DMEM medium containing 5% FBS, 1 ng/ml TGFβ1, 5 ng/ml PDGF-BB, 5 ng/ml bFGF and 5 ng/ml EGF. VECs were purified by sorting of double positive CD31+ /CD144+ population. The sorted cells were cultured on fibronectin coated plate in VECs maintaining medium (F12K basal medium, 20 ng/ml VEGF-A, 20 ng/ml IGF, 5 ng/ml bFGF, 5 ng/ml EGF, 2 μM Heparin and 1X ITS). iPSCs differentiated NSCs, VSMCs and VECs were also characterized using flow cytometry with corresponding antibodies (NESTIN and SOX2 for NSCs, αSMA for VSMCs and CD31 for VECs).
MSCs derived from human iPSCs and related assays
MSCs were differentiated as previously described36. Briefly, iPSCs were firstly induced to neural ectoderm in medium (50% Neurobasal, 50% DMEM/F12 medium, 1% N2 and 2% B27 and 3–6 μM CHIR-99021 and 10 μM SB-431542) for 7 days and switched to MSCs medium (alpha- MEM basal medium, 10% FBS, 1X Glutamax solution, 5 ng/ml bFGF, 5 ng/ml EGF and 1X Penicillin-Streptomycin) for another 7 days. Split the cell at low ratio, such as 1:2–1:3 at early passages with TrypLE Express. MSCs surface markers were characterized using fluorescent antibodies (antibodies from BD 562245 and Biolegend) at passage 5. For adipocyte differentiation, cells at passage 5 growing to 90–100% confluency were washed with 1X PBS for three times before switched to adipocyte induction medium, consisting of HG-DMEM basal medium supplemented with 10% FBS, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 100 µM Indomethacin, 1 µM Dexamethasone, 10 µg/ml Insulin and 1X Penicillin-Streptomycin solution, with medium change every 3 days. After 21 days of induction, cells were fixed in 4% paraformaldehyde and stained with Oil Red O solution. For osteoblast differentiation, cells at passage 5 growing to 90–100% confluency were washed with 1X PBS for three times before switched to osteoblast induction medium, consisting of HG-DMEM basal medium supplemented with 10% FBS, 50 μg/ml L-Ascorbic acid, 0.1 µM Dexamethasone, 10 mM β-glycerophosphate, 1 mM Sodium pyruvate and 1X Penicillin-Streptomycin solution, with medium change every 3 days. After 21 days of induction, cells were fixed in 4% paraformaldehyde and stained with Alizarin Red S solution. For chondrocyte differentiation, cells at passage 5 growing to 90–100% confluency were washed with 1X PBS for three times before switched to chondrocyte induction medium, consisting of LG-DMEM basal medium supplemented with 10% FBS, 50 μg/ml L-Ascorbic acid, 0.1 µM Dexamethasone, 10 ng/ml TGFβ3, 1 mM Sodium pyruvate, 1X Insulin-Transferrin-Selenium solution and 1X Penicillin-Streptomycin solution, with medium change every 3 days. After 21 days of induction, cells were fixed in 4% paraformaldehyde and stained with Alcian blue solution.
Senescence β-Galactosidase Staining
SA-b-gal staining was performed using the commercial kit (Beyotime Biotechnology) according to instruction manual. In brief, cells were fixed in fixative at RT for 5 min. After fixation, cells were stained with freshly prepared staining solution at 37°C overnight. Images were taken and the percentage of senescent cells were analyzed by ImageJ for quantitative analysis.
Immunostaining
The cells were blocked with blocking buffer (PBST containing 5% FBS and 5% BSA) for 1 hour after fix with 4% polyformaldehyde and permeabilization before incubated with primary antibodies at 4 °C overnight. After washing with PBST, the cells were incubated in secondary fluorescent antibodies for 1 hour and counterstained with DAPI (Life Technology). Images were acquired with the confocal microscope with z-stacks (Carl Zeiss LSM800) and were processed using ZEN Blue software with maximum projections. All antibodies source and working concentration can be found in the Supplementary Data 2.
Reverse-transcription PCR and quantitative PCR
RNA was extracted from culture cells by Trizol (Sigma) and 1 μg RNA was used for cDNA synthesis. For reverse PCR, cDNAs were diluted to 200-fold as template and high-fidelity DNA polymerase was used. For qPCR, SYBR Green PCR Kit (Applied Biosystems) was used with cDNAs diluted 50-fold as template and PCR was carried out at standard thermal cycling conditions (95 °C for 20 s; 40 cycles 95 °C for 20 s; 60°C for 30 s. For melting curve, 95 °C for 15 s; 60°C for 1 minute; 95 °C for 15 s), three different repeats were performed and ΔΔCT values were calculated for statistical analysis. The detailed information of primer sequences is list in Supplementary Data 1.
ATAC-Seq
ATAC-seq assay was performed with 5 × 104 beginning human MSCs as previously described81. Human MSCs were lysed with Hypotonic Buffer (10 mM Tris-Cl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% v/v NP-40) on ice for 10 mins and pelleted by centrifugation at 1000 g for 5 min. Transposition reactions were prepared by re-suspending nucleus with 38 μl ddH2O, 10 μl TAPS-MgCl2-DMF buffer and 2 μl assembled Tn5 transposome (5 μM, homemade), followed by incubation at 37 °C for 30 mins. The tagmentated genome DNA purified by DNA purification Kit was eluted with 20 μl TE buffer. ATAC-seq library was set up with 25 μl of NEBNext Ultra II Q5 master mix, 4 μl of 5 mM i5-index and i7-index primer mixture, 5 μl of purified DNA and 16 μl ddH2O. PCR reaction was carried out at 72 °C for 5 min firstly, 98 °C 30 s for denaturation, followed by 11 cycles of 98 °C for 10 s, 63 °C for 30 s and 72 °C for 30 s, and additional 72 °C 5 min for elongation. The ATAC-seq libraries were pooled together and purified with 1.2 x AMPure XP beads before next generation sequencing with paired-end 150-bp reads on NovaSeq 6000 Sequencing System (Novogene Co., Ltd.).
Chromatin immunoprecipitation (ChIP) and ChIP-seq library preparation
Briefly, about 5 106 MSCs were collected, fixed with 1% formaldehydes, quenched with glycine, washed twice with PBS and transferred to −80 °C after snap-frozen. Cells were suspended in 1 mL hypotonic buffer to isolate nucleus, and subsequently lysed with 200 μl nuclei lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.0, EDTA-free protease inhibitor cocktails, 1 mM PSMF and 10 mM sodium pyruvate). The lysates were further sonicated with Diagenode Bioruptor Pico (HKU, Imaging & Flow Cytometry Core facility) for 15–20 cycles to generate chromatin fragments ranging from 200–1000 bp. The chromatin supernatants were further diluted with 800 μl ChIP dilution buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 200 mM NaCl) and incubated with 5 μg antibody at 4 °C overnight. The chromatin-antibody mixture was then incubated with 30 μl blocked protein A or protein G magnetic Dynabeads (Thermofisher) for 3 h. Afterwards, the immunoprecipitated chromatin was washed with three times low salt washing buffer (0.1% SDS,1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0,150 mM NaCl), three times high salt washing buffer (0.1% SDS,1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 500 mM NaCl) and once LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% Sodium Deoxycholate,1 mM EDTA, 10 mM Tris-HCl pH 8.0) sequentially before subjected to proteinase K digestion. The ChIP DNAs were extracted by phenol-chloroform-isoamyl alcohol method and quantified by qubit fluorometer (Thermofisher). ChIP libraries were prepared by NEBNext® ΜltraTM DNA Library Prep Kit using Illumina® (NEB E7370) following the instruction manual. The deep sequencing was performed with paired-end 150-bp reads on the NovaSeq 6000 Sequencing System (Novogene Co., Ltd.).
Bridge linker based Hi-C (BAT-Hi-C)
BAT-Hi-C was performed according to a previously published protocol82. Cells (5 × 106 – 1 × 107) were prepared as above mentioned and lysed in 500 μl of lysis buffer ((0.1%SDS, 50 mM HEPES-KOH, pH7.5; 150 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% Sodium Deoxycholate) containing protease inhibitors, and incubated on ice for 20 min. Cells were washed twice in lysis buffer and incubated in 50 μl of 0.5% SDS at 62 °C for 10 min. The SDS was quenched with 25 μl of 10% Triton-X and 145 μl H2O at 37 °C for 15 min. NEB Buffer 2 and 100 U of AluI (NEB) were added and the chromatin was digested at 37 °C overnight with gentle rotation. Nuclei were collected after AluI inactivation. Resuspended in 400 μl of Klenow (3’−5’exo-) solution (40 μl NEB Buffer 2, 8 μl 10 mM dATP, 40 μl 10% TritonX-100, 304 μl H2O, and 8 μl Klenow (3’−5’exo-)) and incubated at 37 °C for 1 h. Washed nuclei twice with 1× T4 DNA ligase buffer followed by resuspending in 1200 μl of proximity ligation solution (120 μl T4 DNA ligase buffer, 120 μl 10% TritonX-100, 939 μl H2O, 6 μl T4 DNA ligase, 12 μl 10 mg/ml BSA, 3 μl Bridge linker (200 ng/μl)) and rotating at ~22°C for 4 h. Centrifuged at 3500 g for 5 min at 4 °C to remove the supernatant, and then added 70 μl lambda exonuclease buffer, 4 μl lambda exonuclease (NEB), 6 μl exonuclease I (NEB), 618 μl H2O, rotated at 37 °C for 1 h. For reverse crosslinking, chromatin was resuspended with 1370 μl digestion buffer (10 mM Tris-HCl, pH 8, 25 mM EDTA, 1% SDS, 1 mg/ml proteinase K), incubated at 55 °C for 1 h. Then, 130 μl 5 M NaCl was added and incubated at 68 °C overnight with shaking. DNA was purified by phenol chloroform extraction and ethanol precipitation, followed by sonication using Covaris Focused-ultra-sonicator to 200–1000 bp. The biotinylated DNA fragments were further enriched using 30 μl of 10 mg/ml Dynabeads MyOne Streptavidin T1 beads. Hi-C libraries were prepared using NEBNext® ΜltraTM DNA Library Prep Kit and amplified with 10–12 PCR cycles. The final products were size-selected with AMPure XP beads and sequenced with paired-end 150-bp reads on the NovaSeq 6000 Sequencing System (Novogene Co., Ltd.).
Quality control of sequencing reads
All the Illumina sequencing reads used in the study were firstly quality controlled by Trim Galore (version 0.6.7, https://github.com/FelixKrueger/TrimGalore). In details, we removed the bases with quality below 20 and the adapter sequences from the 3’ end and filtered the reads with length less than 50nt.
RNA-seq analysis
For read alignment and expression quantification, we first removed low quality reads, and trimmed the adapter sequence with Trim Galore (version 0.6.7, https://github.com/FelixKrueger/TrimGalore). Then we mapped the remaining pair-end reads to the reference genome hg19 using STAR83 with ENCODE option bundles. Using HTSeq-count84, we counted the uniquely mapped reads, and normalized the read count by trimmed mean of M values (TMM), and transformed to reads per kilobases per million reads (RPKM) by edgeR85. With an expression cutoff of RPKM ≥ 1 in at least one sample group, we removed low abundant genes, and detected the differentially expressed genes using edgeR. Genes were considered differentially expressed when the overall false discovery rate (FDR) < 0.05 and fold change is above 2.0.
Gene ontology (GO) analysis
Gene symbols were first converted to EntrzID with R package BiomaRt86 (version 2.42.0). ‘ensembl’ was used as biomart database, and ‘mmusculus_gene_ensembl’ was used as dataset. EntrzID of interest genes was uploaded to DAVID 6.887 (https://david.ncifcrf.gov).
Comparison with published human MSCs aging models
The published transcriptome data of hMSCs aging models were downloaded and re-processed from GEO GSE26093 (labeled as “HGPS”, differentially expressed genes were calculated by comparing HGPS-iPS-MSC to N-iPS-MSC with detection p-value of probe <0.05 and absolute value of fold change > 2.0), GSE39540 (labeled as “AgeCorrelate”, differentially expressed genes were reported in Table 2 of the reference paper)61, GSE35958 (differentially expressed genes labeled as “NomalOld” were reported in Table S2 column 3 and “NormalSenecent” were reported in Table S2 column 4 of the reference paper)88, GSE39250 (labeled as “TERT_OE”, differentially expressed genes were from Table S1 of the reference paper with q-value% <12 as reported)89, GSE48662 (labeled as “Irradiation”, differentially expressed genes were reported in Table S3 and Table S4 of the reference paper)90, GSE52285 (labeled as “WRN_KO”, differentially expressed genes were reported in Table S2 Sheet 3 and 4 of the reference paper)30, GSE64642 (labeled as “SIRT6_KO”, differentially expressed genes were reported in Table S2 of the reference paper)91, GSE84694 (labeled as “NRF2_mut”, differentially expressed genes were calculated by comparing AG_MSC_LP to WT_MSC_LP with FPKM ≥ 1 of both replicates in at least one sample group and absolute value of log2 fold change > 0.58)92, GSE102004 (labeled as “ATF6_KO”, differentially expressed genes were reported in Table S3 of the reference paper)93, GSE122654 (labeled as “RAP1_KO”, differentially expressed genes were calculated by HTSeq-count and edgeR as described in “RNA-seq analysis” part with RPKM ≥ 1 of both replicates in at least one sample group and absolute value of log2 fold change > 0.58)94, GSE117084 (labeled as “CBX4_KO”, differentially expressed genes were reported in Table S1 Sheet 3 and 4 of the reference paper)95, GSE145019 (labeled as “CLOCK_KO”, differentially expressed genes were reported in Table S4 of the reference paper)96, GSE116277 (labeled as “FOXO3_mut”, differentially expressed genes were reported in Table S2 Sheet 7 and 8 of the reference paper)97, GSE113117 (labeled as “DGCR8_mut”, differentially expressed genes were calculated by HTSeq-count and edgeR as described in “RNA-seq analysis” part with RPKM ≥ 1 of both replicates in at least one sample group and absolute value of log2 fold change > 0.58)98, GSE110268 (differentially expressed genes labeled as “YAP_KO” were reported in S2 Data column 1–15 and “FOXD1_KO” were reported in S5 Data of the reference paper)99, GSE146387 (labeled as “ZKSCAN3_KO”, differentially expressed genes were reported in Table S3 Sheet 1 and 2 of the reference paper)100, GSE146247 (labeled as “SIRT7_KO”, differentially expressed genes were reported in Supplementary material 5 of the reference paper)101, GSA HRA000466 (labeled as “SIRT3_KO”, differentially expressed genes were reported in Table S4 Sheet 2 of the reference paper)102, GEO GSE175733 (differentially expressed genes labeled as “GAtreatHGPS” and “GAtreatWS” were respectively calculated by HTSeq-count and edgeR as described in “RNA-seq analysis” part with RPKM ≥ 1 of both replicates in at least one sample group and absolute value of log2 fold change > 0.58, genes with top 100 average RPKM in the three replicates of “DMSO_HGPS” were labeled as “HGPS_DMSO” and genes with top 100 average RPKM in the three replicates of “DMSO_WS” were labeled as “WS_DMSO”. “GAtreat” means that the cells were treated by gallic acid (GA), which is a natural phenolic compound with antioxidant, anti-inflammatory, and antineoplastic properties)103 and GSE124197 (genes labeled as “SenecenceDriver” were reported in Data file S1 Sheet 1 and differentially expressed genes labeled as “WS” were reported in Data file S1 Sheet 3 of the reference paper)104.
As listed in Extended Data Fig. 5a, those human MSCs aging models were classified into “Accelerated hMSCs” and “Alleviated hMSCs” according to published cellular phenotypes. The up-regulated genes in Accelerated hMSCs, down-regulated genes in Alleviated hMSCs, top 100 genes in “HGPS_DMSO” and “WS_DMSO”, and “SenesenceDriver” were defined as “senescence-associated genes” in this study, while the down-regulated genes in Accelerated hMSCs and up-regulated genes in Alleviated hMSCs were defined as “geroprotection-associated genes”. The relationship between those gene lists were quantified by hypergeometric tests in Figs. 3j, 4g, 5e, and 7k. The hypergeometric tests were performed by homemade Perl script which call “phyer” function in R with the parameter “lower.tail = FALSE” and adjusted the p-value by “p.adjust” function with “method = ‘fdr’”. Clustering of the results of hypergeometric test was performed and visualized by pheatmap using ward’s method (version 1.0.12, https://cran.r-project.org/web/packages/pheatmap).
ChIP-seq and ATAC-seq analysis
Fastq files were trimmed adapters by Trim Galore (version 0.6.7, https://github.com/FelixKrueger/TrimGalore) and aligned to hg19 reference genome using Bowtie2 (version 2.2.5, http://bowtie-bio.sourceforge.net/bowtie2/) with default parameters. Reads with a mapq score less than 30 and PCR duplications were filtered out by using Samtools105 (version 1.9, http://samtools.sourceforge.net). Reads aligned to the regions in ENCODE blacklist (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/) were discarded through bedtools106 (version 2.29.1, https://bedtools.readthedocs.io/en/latest/). ATAC-seq data were processed same as ChIP-seq data. Peaks were called with MACS2107 (version 2.1.2, parameters: ‘-q 0.05 -m 5 50’ for ATAC-seq, H3K27ac and CTCF, ‘-q 0.05 -m 5 50 --broad’ for H3K27me3, H3K9me3, Lamin A/C and Lamin B1) using input as control.
LAD and non-LAD lamina-chromatin binding peaks analysis
The replicates of Lamin A/C and Lamin B1 ChIP and input BAM files were combined and down-sampled to 16 M read counts prior to LAD calling using Samtools105 (version 1.9, http://samtools.sourceforge.net). LADs were called using Enriched Domain Detector47 (EDD, version 1.1.19, http://github.com/CollasLab/edd, parameters: ‘max_CI_value = 0.25, required_fraction_of_informative_bins = 0.80, p_hat_CI_method = agresti_coull, log_ratio_bin_size = 10’). The non-LAD lamin-chromatin binding sites were Lamin peaks out of LADs, calculated by bedtools106 (intersectBed -v). In details, the calling of LADs or peaks were performed on each replicate before calling the consensus LADs or peaks. Lamin peaks were called with MACS2 with parameters detailed in ‘ChIP-seq and ATAC-seq analysis—Data processing’ section.
Enrichment analysis
Enrichment analysis of ChIP on different regions in Figs. 4c, 6c–f, h, i, k, l, and 7j were performed by deepTools108 (version 3.1.1, https://deeptools.readthedocs.io/en/develop/). In details, BAM files were converted to bigwig files using “bamCoverage” function with normalization method “RPKM” and compared to Input by “bigwigCompare” function with default parameters. The signals in different regions were calculated by “computeMatrix” function with parameters ‘-m 1 -b 0 -a 0 -bs 1 --averageTypeBins mean’ and visualized by R package ggplot2 (version 3.3.5, https://cran.r-project.org/web/packages/ggplot2/index.html). All p-values were determined by R package ggpubr (version 0.4.0, https://cran.r-project.org/web/packages/ggpubr/index.html) with Wilcoxon rank-sum test.
Differential analysis
Differential regions of LADs were computed by “subtractBed” function of bedtools (version 2.29.1, https://bedtools.readthedocs.io/en/latest/). Differential analysis of ChIP were performed by R package DiffBind (version 3.2.5, https://bioconductor.org/packages/release/bioc/html/DiffBind.html, parameters: ‘dba.count(summits = 250), dba.contrast(minMembers = 2) and dba.analysze() with default parameters’). Peaks were considered differential when FDR < 0.05. RPKM of differential peaks were converted to z-score and visualized by pheatmap (version 1.0.12, https://cran.r-project.org/web/packages/pheatmap).
Permutation test of overlapped regions
Permutation test in Fig. 5c was performed by homemade Python3 script. In details, numbers of lamin-peaks out of LADs overlapping with gene promoters (TSS ± 3 kb) were compared to a null distribution of numbers consisting of 1000 draws of randomly shuffled Lamin-peaks overlapping with gene promoters. The overlapping was performed by bedtools and all p-value of permutation test were determined by R function “pnorm”.
Peak density analysis
Density of differential CTCF peaks in different regions in Fig. 7h were calculated by numbers of CTCF peaks within interested regions divided by total length of the regions and normalized to the ratio of this value to the value of all CTCF peaks in the whole genome.
Hi-C analysis
Adapters were first trimmed by Trim Galore (version 0.6.7, https://github.com/FelixKrueger/TrimGalore). Next, bridge linkers of paired-end reads were trimmed with trimLinker program (part of ChIA-PET2, version 0.9.3, https://github.com/GuipengLi/ChIA-PET2, parameters: ‘-t 20 -m 1 -k 1’). Subsequently, the Hi-C paired-end reads were aligned to the hg19 reference genome and paired using HiC-Pro109 (version 2.11.1, parameters: ‘LIGATION_SITE = AGCT’ which digested hg19 by AluI). The correlation of Hi-C matrices was analyzed by hicCorrelate (part of hicexplorer110, version 3.5.3, parameter: ‘--range 5000:200000, --method=pearson, --log1p’). The two replicates were merged in the following analysis and visualization (except for differential domain boundaries detection, details in ‘TAD analysis’). Then the data was used to generate contact metrices and corrected with ice_norm111 (part of HiC-Pro). For each chromosome, the ICE normalized 10 kb, 40 kb and 100 kb resolution contact matrices were used for further analysis. The 40 kb ICE normalized contact metrices of each chromosome were used to analyze relative contact probability (RCP) with GENOVA (version 0.9, https://github.com/robinweide/GENOVA).
For compartmental analysis, the first eigenvector (PC1) values were calculated from ICE normalized matrices (100 kb bin) at each chromosome separately, with CscoreTool112 (version 1.1, parameters: 20 1000000). Next, each bin was assigned into A or B compartments according to its PC1 values and average gene density across the whole regions by homemade Perl script. The gene-rich compartments were finally defined as A, and the gene-poor compartments as B. For compartment display, the absolute values of PC1 were kept, positive value represents compartment A, whereas negative value represents compartment B. Finally, considering a TAD is always contained in one compartment, the compartments were smoothed in TAD level. TADs were assigned to either the A- or the B- compartments, by calculating the average dominant eigenvector of each TAD. Compartment regions were determined by combining adjacent bins with same type. The 100 kb resolution contact matrices were transformed into an observed over expected (O/E) matrices with hicTransform110 (part of hicexplorer, version 3.5.3, parameter: ‘--method obs_exp_lieberman’) before calculating average contacts. Quantification of average A-A, B-B, A-B contacts and visualization of saddle plots in Fig. 7b were done by cooltools compute-saddle (version 0.3.2, https://github.com/open2c/cooltools, parameter: ‘-n 50 --qrange 0.005 0.995’) using 100 kb ICE normalized O/E matrices. Compartment strength was calculated by average A-A or B-B contacts divided by average A-B contacts. The homotypic interaction saddle plots in Fig. 7c were also drawn by cooltools compute-saddle, but with bins sorted based on their signal of Lamin bindings rather than PC1 as previous reported113.TAD boundaries were detected using 40 kb ICE-normalized matrices by TopDom54 (version 0.0.2, http://zhoulab.usc.edu/TopDom, parameters ‘w = 5’), a continuous quantitative method that has been proved to be robust to resolution and sequence depth114. The differential TAD boundaries using diffHic115 (version 1.22.0, https://anaconda.org/bioconda/bioconductor-diffhic). In details, the valid HiC interaction reads were converted to HD5 files by each normalized replicate, and used to calculate the Direction Index by “domainDirections” function setting width as 40k and span as 7. The significant differences test with replicates were performed by “glmQLFTest” function. Only the raw TopDom TAD boundaries with FDR < 0.05 were treated as reorganization, and their corresponding TAD were classified as shortened, unchanged length and enlarged by homemade Perl script. Chromatin loops of Hi-C were determined by cLoops116 (v0.93, https://github.com/YaqiangCao/cLoops, parameters: ‘-eps 10000 -minPts 5 -hic’). The loops were considered significant when the “significant” column (15th) of cLoops outputs equals 1. Differential analysis of loops were performed by diffloop117 (v1.9.0, https://github.com/aryeelab/diffloop, “quickAssoc”) and E-P loops were selected by “keepEPloops” with the union set of WT and MAD enhancers. Only the E-P loops with FDR < 0.05 were considered as altered loops. APA analysis was performed with 10 kb ICE normalized matrices using GENOVA (version 0.9, https://github.com/robinweide/GENOVA) with default parameters in Fig. 7a. Genomic browser view of ChIP-seq, ATAC-seq, and RNA-seq tracks in Figs. 4a, 5a, 6a,and 7j were plotted using IGV118 (version 2.4.10). Pearson correlation matrices in Fig. 7a and contact matrices in Fig. 7j were visualized using Juicebox119 (version 1.11.08, https://www.aidenlab.org/juicebox). Figure 2a, Extended Data Fig. 4a, d and, g contained modified images from Servier Medical Art (https://smart.servier.com) licenced by a Creative Commons Attribution 4.0 Unported License.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank Dr Daniel Mak for proofreading the manuscript. Z.Z. was supported by grants from InnoHK@Health, Theme-based Research Scheme (Tl3-602/21-N), Guangdong High-level Hospital Construction Project (KJ012019517) and Guangdong Provincial People’s Hospital Foundation (KY012021405), Guangdong-Dongguan Joint Research Scheme Guangdong-Hong Kong-Macau Program (2021B1515130004), Guangdong Innovative and Entrepreneurial Research Team Program (2016ZT06S029), Guangdong Basic and Applied Basic Research Foundation (2021B1515120063, 2021B1515120063), Shanghai Pujiang Program (2020PJD063), and National Natural Science and Foundation of China (81500447, 81671399, 81971329, 31771639, 31970811, 32425022 and 32170798). T. L. was supported by The Hong Kong Scholars Program (XJ2016016). J.D. was supported by the National Key R&D Program of China (2023YFA1800900).
Author contributions
J.W. and Z.Z. conceived the project and designed the experiments. J.W. did all the primary cell isolation, iPSCs generation and differentiation, and ChIP Assays. S.J. performed the Hi-C experiment. XY. Liu. and J.D. analyzed the sequencing data. Y. H., T.L. performed the experiments on cellular senescence, DNA damage analyzes, and teratoma formation. J.M. and X. Lu. helped to prepare dermal fibroblasts from human patients. Z.C., XG. Liu, W.S., and G.J. helped with data analysis and discussion. J.W. and Z.Z. interpreted the data and prepared the manuscript with comments and inputs from all authors.
Peer review
Peer review information
Nature Communications thanks Chiara Lanzuolo, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The RNA-seq, ATAC-seq, Hi-C and ChIP-seq data generated in this study have been deposited in the GEO database under accession code GSE193694. The differential gene lists and all chromatin loops generated in this study are provided in the Supplementary Information. The published transcriptome data of hMSCs aging models are listed in the following: GSE26093 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE26093], GSE39540 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE39540], GSE35958, GSE39250, GSE48662, GSE52285, GSE64642, GSE84694, GSE102004, GSE122654, GSE117084, GSE145019, GSE116277, GSE113117, GSE110268, GSE146387, GSE146247, GSA HRA000466 GSA HRA000466 [10.1093/nar/gkab161], GSE175733 and GSE124197. Source data are provided in this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wei Jin, Shaoshuai Jiang, Xinyi Liu, Yi He.
Contributor Information
Junjun Ding, Email: dingjunj@mail.sysu.edu.cn.
Zhongjun Zhou, Email: zhongjun@hku.hk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54338-3.
References
- 1.Rankin, J. & Ellard, S. The laminopathies: a clinical review. Clin. Genet70, 261–274 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Doubaj, Y. et al. An inherited LMNA gene mutation in atypical Progeria syndrome. Am. J. Med Genet A158A, 2881–2887 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Csoka, A. B. et al. Novel lamin A/C gene (LMNA) mutations in atypical progeroid syndromes. J. Med Genet. 41, 304–308 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg, A. et al. Atypical progeroid syndrome due to heterozygous missense LMNA mutations. J. Clin. Endocrinol. Metab.94, 4971–4983 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L. et al. LMNA mutations in atypical Werner’s syndrome. Lancet362, 440–445 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Cenni, V. et al. Mandibuloacral dysplasia: a premature aging disease with aspects of physiological aging. Ageing Res Rev.42, 1–13 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Liu, G. H. et al. Recapitulation of premature aging with iPSCs from Hutchinson-Gilford progeria syndrome. Nature472, 221–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, H., Xiong, Z. M. & Cao, K. Mechanisms controlling the smooth muscle cell death in progeria via down-regulation of poly(ADP-ribose) polymerase 1. Proc. Natl Acad. Sci. USA111, E2261–E2270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood, A. M. et al. TRF2 and lamin A/C interact to facilitate the functional organization of chromosome ends. Nat. Commun.5, 5467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, B. et al. Resveratrol rescues SIRT1-dependent adult stem cell decline and alleviates progeroid features in laminopathy-based progeria. Cell Metab.16, 738–750 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Ghosh, S., Liu, B., Wang, Y., Hao, Q. & Zhou, Z. Lamin A Is an endogenous SIRT6 activator and promotes SIRT6-mediated DNA repair. Cell Rep.13, 1396–1406 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Pegoraro, G. et al. Ageing-related chromatin defects through loss of the NURD complex. Nat. Cell Biol.11, 1261–U251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, B. et al. Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a progeria mouse model. Nat. Commun.4, 1868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattioli, E., Andrenacci, D., Garofalo, C. & Cicchillitti, L. Altered modulation of lamin A/C-HDAC2 interaction and p21 expression during oxidative stress response in HGPS (vol 17, e12824, 2018). Aging Cell17, e12824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lionetti, M. C. et al. Chromatin and cytoskeletal tethering determine nuclear morphology in progerin-expressing cells. Biophys. J.118, 2319–2332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebestyen, E. et al. SAMMY-seq reveals early alteration of heterochromatin and deregulation of bivalent genes in Hutchinson-Gilford Progeria Syndrome. Nat. Commun.11, 6274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong, Z. M. et al. Methylene blue alleviates nuclear and mitochondrial abnormalities in progeria. Aging Cell15, 279–290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubben, N. et al. Repression of the antioxidant NRF2 pathway in premature aging. Cell165, 1361–1374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briand, N. & Collas, P. Lamina-associated domains: peripheral matters and internal affairs. Genome Biol.21, 85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J. et al. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature572, 335–340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheedipudi, S. M. et al. Genomic reorganization of lamin-associated domains in cardiac myocytes is associated with differential gene expression and DNA methylation in human dilated cardiomyopathy. Circ. Res. 124, 1198–1213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salvarani, N. et al. The K219T-Lamin mutation induces conduction defects through epigenetic inhibition of SCN5A in human cardiac laminopathy. Nat. Commun.10, 2267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCord, R. P. et al. Correlated alterations in genome organization, histone methylation, and DNA-Iamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res.23, 260–269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falk, M. et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature570, 395–399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang, J. et al. ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat. Commun.9, 1364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ucar, D. et al. The chromatin accessibility signature of human immune aging stems from CD8(+) T cells. J. Exp. Med. 214, 3123–3144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen, P. et al. Histone acetyltransferase p300 induces de novo super-enhancers to drive cellular senescence. Mol. Cell73, 684–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasdemir, N. et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov.6, 612–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan, Y. T. et al. Senescence-activated enhancer landscape orchestrates the senescence-associated secretory phenotype in murine fibroblasts. Nucleic Acids Res.48, 10909–10923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, W. et al. Aging stem cells. a werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science348, 1160–1163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nativio, R. et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat. Neurosci.21, 497–505 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sati, S. et al. 4D Genome rewiring during oncogene-induced and replicative senescence. Mol. Cell78, 522–538 e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Criscione, S. W. et al. Reorganization of chromosome architecture in replicative cellular senescence. Sci. Adv.2, e1500882 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandra, T. et al. Global reorganization of the nuclear landscape in senescent cells. Cell Rep.10, 471–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zirkel, A. et al. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol. Cell70, 730–73 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Jin, W. et al. Rapid and robust derivation of mesenchymal stem cells from human pluripotent stem cells via temporal induction of neuralized ectoderm. Cell Biosci.12, 31 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, D. Q. et al. Mandibuloacral dysplasia type A-associated progeria caused by homozygous LMNA mutation in a family from Southern China. Bmc Pediatrics14, 256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh, J., Lee, Y. D. & Wagers, A. J. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat. Med. 20, 870–880 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Signer, R. A. J. & Morrison, S. J. Mechanisms that Regulate Stem Cell Aging and Life Span. Cell Stem Cell12, 152–165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov, A. et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol.202, 129–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature527, 105–109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah, P. P. et al. Pathogenic LMNA variants disrupt cardiac lamina-chromatin interactions and de-repress alternative fate genes. Cell Stem Cell28, 938–954 e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lund, E. et al. Lamin A/C-promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res.23, 1580–1589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimi, T. et al. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev.25, 2579–2593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah, P. P. et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev.27, 1787–1799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lund, E., Oldenburg, A. R. & Collas, P. Enriched domain detector: a program for detection of wide genomic enrichment domains robust against local variations. Nucleic Acids Res. 42, e92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Giorgio, E. et al. HDAC4 degradation during senescence unleashes an epigenetic program driven by AP-1/p300 at selected enhancers and super-enhancers. Genome Biol.22, 129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barone, G. et al. Human CDK18 promotes replication stress signaling and genome stability. Nucleic Acids Res. 44, 8772–8785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vance, K. W., Carreira, S., Brosch, G. & Goding, C. R. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res.65, 2260–2268 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Borbolis, F. et al. mRNA decapping is an evolutionarily conserved modulator of neuroendocrine signaling that controls development and aging. Elife9, e53757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borbolis, F. & Syntichaki, P. Cytoplasmic mRNA turnover and aging. Mech. Ageing Dev.152, 32–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohler, F. et al. Epigenetic deregulation of lamina-associated domains in Hutchinson-Gilford progeria syndrome. Genome Med. 12, 46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin, H. J. et al. TopDom: an efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res.44, e70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giorgetti, L. et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell157, 950–963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levi, N., Papismadov, N., Solomonov, I., Sagi, I. & Krizhanovsky, V. The ECM path of senescence in aging: components and modifiers. FEBS J.287, 2636–2646 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Mays, P. K., McAnulty, R. J., Campa, J. S. & Laurent, G. J. Age-related changes in collagen synthesis and degradation in rat tissues. Importance of degradation of newly synthesized collagen in regulating collagen production. Biochem J.276, 307–313 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tominaga, K. & Suzuki, H. I. TGF-beta signaling in cellular senescence and aging-related pathology. Int J. Mol. Sci.20, 5002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gil, J., Bernard, D., Martinez, D. & Beach, D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell Biol.6, 67-U19 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Malaquin, N. et al. Senescent fibroblasts enhance early skin carcinogenic events via a paracrine MMP-PAR-1 axis. PLoS One8, e63607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, J. Q. et al. A human iPSC model of Hutchinson Gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell8, 31–45 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Ocampo, A. et al. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell167, 1719–1733 e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nissan, X. et al. Unique preservation of neural cells in Hutchinson–Gilford progeria syndrome is due to the expression of the neural-specific miR-9 microRNA. Cell Rep.2, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Jung, H. J. et al. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl Acad. Sci. USA109, E423–E431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guelen, L. et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature453, 948–951 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Kind, J. et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell163, 134–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng, X. et al. Lamins organize the global three-dimensional genome from the nuclear periphery. Mol. Cell71, 802–815 e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leemans, C. et al. Promoter-intrinsic and local chromatin features determine gene repression in LADs. Cell177, 852–864 e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pascual-Reguant, L. et al. Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat. Commun.9, 3420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ikegami, K., Secchia, S., Almakki, O., Lieb, J. D. & Moskowitz, I. P. Phosphorylated lamin A/C in the nuclear interior binds active enhancers associated with abnormal transcription in progeria. Dev. Cell52, 699–713 e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, F. & Yao, J. Identifying novel transcriptional and epigenetic features of nuclear lamina-associated genes. Sci. Rep.7, 100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amendola, M. & van Steensel, B. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep.16, 610–617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lenain, C., Gusyatiner, O., Douma, S., van den Broek, B. & Peeper, D. S. Autophagy-mediated degradation of nuclear envelope proteins during oncogene-induced senescence. Carcinogenesis36, 1263–1274 (2015). [DOI] [PubMed] [Google Scholar]
- 74.Xu, C. et al. SIRT1 is downregulated by autophagy in senescence and aging. Nat. Cell Biol.22, 1170–1179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu, Z. et al. Large-scale chromatin reorganization reactivates placenta-specific genes that drive cellular aging. Dev. Cell57, 1347–1368.e12 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Yang, Y. K., Ogando, C. R., Wang See, C., Chang, T. Y. & Barabino, G. A. Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Res Ther.9, 131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pittenger, M. F. et al. Mesenchymal stem cell perspective: cell biology to clinical progress. NPJ Regen. Med. 4, 22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mastrolia, I. et al. Challenges in clinical development of mesenchymal stromal/stem cells: concise review. Stem Cells Transl. Med. 8, 1135–1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Musial-Wysocka, A., Kot, M. & Majka, M. The pros and cons of mesenchymal stem cell-based therapies. Cell Transpl.28, 801–812 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patsch, C. et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol.17, 994–1003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liang, Z. et al. BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions. Nat. Commun.8, 1622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smedley, D. et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 43, W589–W598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc.4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 88.Benisch, P. et al. The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS One7, e45142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Estrada, J. C. et al. Human mesenchymal stem cell-replicative senescence and oxidative stress are closely linked to aneuploidy. Cell Death Dis.4, e691 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sepulveda, J. C. et al. Cell senescence abrogates the therapeutic potential of human mesenchymal stem cells in the lethal endotoxemia model. Stem Cells32, 1865–1877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan, H. et al. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res.26, 190–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang, J. et al. Genetic enhancement in cultured human adult stem cells conferred by a single nucleotide recoding. Cell Res. 27, 1178–1181 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang, S. et al. ATF6 safeguards organelle homeostasis and cellular aging in human mesenchymal stem cells. Cell Discov.4, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang, X. et al. Telomere-dependent and telomere-independent roles of RAP1 in regulating human stem cell homeostasis. Protein Cell10, 649–667 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ren, X. et al. Maintenance of Nucleolar Homeostasis by CBX4 Alleviates Senescence and Osteoarthritis. Cell Rep.26, 3643–3656 e7 (2019). [DOI] [PubMed] [Google Scholar]
- 96.Liang, C. et al. Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration. Cell Res. 31, 187–205 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yan, P. et al. FOXO3-engineered human ESC-derived vascular cells promote vascular protection and regeneration. Cell Stem Cell24, 447–461 e8 (2019). [DOI] [PubMed] [Google Scholar]
- 98.Deng, L. et al. Stabilizing heterochromatin by DGCR8 alleviates senescence and osteoarthritis. Nat. Commun.10, 3329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fu, L. et al. Up-regulation of FOXD1 by YAP alleviates senescence and osteoarthritis. PLoS Biol.17, e3000201 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hu, H. et al. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 48, 6001–6018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bi, S. J. et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell11, 483–504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Diao, Z. et al. SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Res. 49, 4203–4219 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shan, H. Z. et al. Large-scale chemical screen identifies Gallic acid as a geroprotector for human stem cells. Protein Cell13, 532–539 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang, W. et al. A genome-wide CRISPR-based screen identifies KAT7 as a driver of cellular senescence. Sci. Transl. Med.13, eabd2655 (2021). [DOI] [PubMed] [Google Scholar]
- 105.Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang, Y. et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol.9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res.42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol.16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wolff, J. et al. Galaxy HiCExplorer 3: a web server for reproducible Hi-C, capture Hi-C and single-cell Hi-C data analysis, quality control and visualization. Nucleic Acids Res.48, W177–W184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods9, 999–1003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zheng, X. B. & Zheng, Y. X. CscoreTool: fast Hi-C compartment analysis at high resolution. Bioinformatics34, 1568–1570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Belaghzal, H. et al. Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Nat. Genet.53, 367–378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dali, R. & Blanchette, M. A critical assessment of topologically associating domain prediction tools. Nucleic Acids Res. 45, 2994–3005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lun, A. T. L. & Smyth, G. K. diffHic: a Bioconductor package to detect differential genomic interactions in Hi-C data. Bmc Bioinforma.16, 258 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cao, Y. et al. Accurate loop calling for 3D genomic data with cLoops. Bioinformatics36, 666–675 (2020). [DOI] [PubMed] [Google Scholar]
- 117.Lareau, C. A. & Aryee, M. J. diffloop: a computational framework for identifying and analyzing differential DNA loops from sequencing data. Bioinformatics34, 672–674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thorvaldsdottir, H., Robinson, J. T. & Mesirov, J. P. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinforma.14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping (vol 159, pg 1665, 2014). Cell162, 687–688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The RNA-seq, ATAC-seq, Hi-C and ChIP-seq data generated in this study have been deposited in the GEO database under accession code GSE193694. The differential gene lists and all chromatin loops generated in this study are provided in the Supplementary Information. The published transcriptome data of hMSCs aging models are listed in the following: GSE26093 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE26093], GSE39540 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE39540], GSE35958, GSE39250, GSE48662, GSE52285, GSE64642, GSE84694, GSE102004, GSE122654, GSE117084, GSE145019, GSE116277, GSE113117, GSE110268, GSE146387, GSE146247, GSA HRA000466 GSA HRA000466 [10.1093/nar/gkab161], GSE175733 and GSE124197. Source data are provided in this paper.