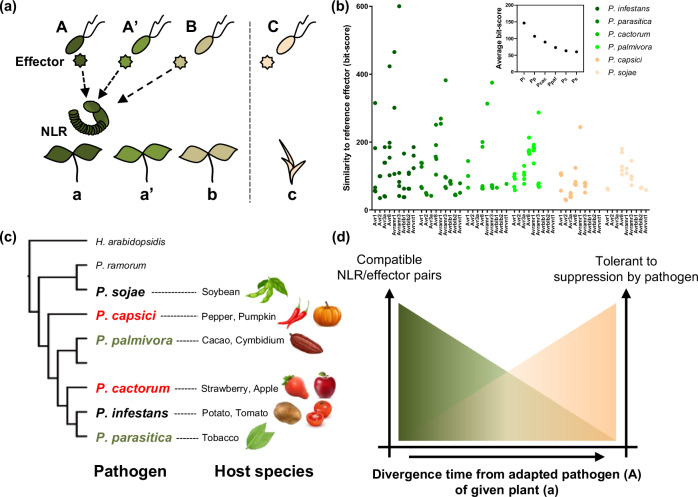
Fig. 5. NLR-mediated recognitions of conserved effectors would contribute to resistance against a wide range of pathogens.
a Pathogens diverged from common ancestral species adapt to its host plant species (with the same color) and this adaptation is accompanied with the changes in repertoire and sequence of effectors. However, effectors are conserved between the recently diverged pathogen species. Thus, a portion of conserved effectors could be recognized by same NLRs. b Bit-scores obtained by comparing homologous effectors (69 cloned effectors, Avramr3 and its homologs are included) against each reference avirulence effector of P. infestans are presented as dot graph, and average bit-score of whole tested effectors of each species is presented in a small boxed graph (above). Conservation of P. infestans reference avirulence effectors across the Phytophthora species gradually decreases with increasing divergence time from P. infestans. c Resistance genes against most Phytophthora species have never been identified (red: no NLRs were reported; green: few NLRs were reported) though they cause serious disease in various crops. d Considering that conservation of effectors decreased with increasing of divergence time between pathogen species, the number of compatible NLR/effector pairs would also similarly decrease. Simultaneously, pathogens are less-likely to suppress NLRs derived from evolutionarily distant plants from its host species.