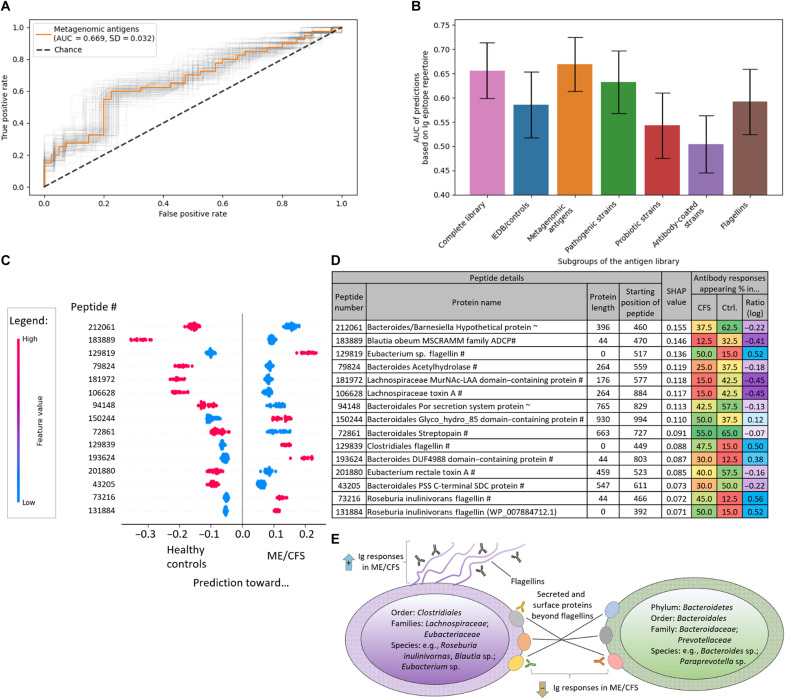
Fig. 3. Machine learning algorithms trained on Ig epitope repertoires separate patients with severe ME/CFS from healthy controls.
The classifications are driven by antibody responses against Lachnospiraceae flagellins overrepresented in patients with severe ME/CFS as well as antibody responses against other surface proteins of Bacteroidetes and Lachnospiraceae in the healthy controls. All predictions were performed with GBR (60) and leave-one-out cross-validation on different prevalence cutoffs of antibody responses appearing in the cohorts (see Methods for details). (A) Receiver operator curve (ROC) of the best-performing combination of antibody-bound peptides and prevalence cutoffs. The light gray lines illustrate ROC confidence intervals (CIs, 95%) as computed by bootstrapping. (B) Summary of model performance trained on antibody responses against different subsets within the antigen library. See Methods for details on the subgroups of the antigen library. Error bars represent 95% CIs as in (A). (C and D) SHAP analysis of the best-performing model [shown in (A)]. The top 15 contributing peptides are shown (C), and details are provided (D). See table S3 for a full list. Every line represents a peptide, and in every line, every dot represents the prediction result contribution for one individual (n = 80; 40 CFS patients and 40 controls). See Methods for abbreviations. (E) Schematic illustration of antibody responses against the respective proteins from Lachnospiraceae, Eubacteriaceae, and Bacteroidetes driving classification.