Figure 4: Intact DNA repair and reduced double strand breaks in HPV+ OPSCCs predisposed to recur.
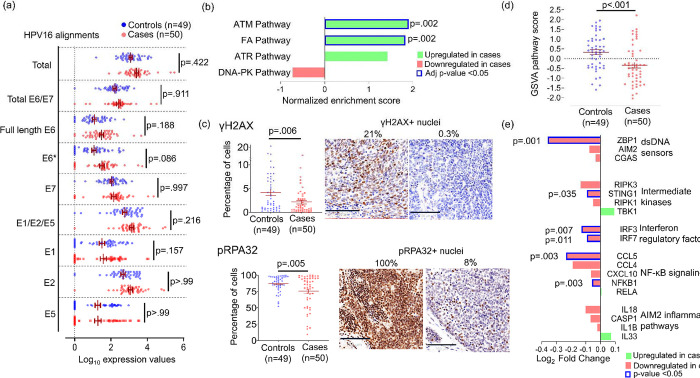
(a) Normalized HPV16 transcript alignments in cases and controls. E6* indicates all spliced E6 forms. Adjusted p-values were calculated using Bonferroni and Sidak’s procedure. (b) DNA repair signaling pathways in the Protein Interaction Database compared between cases and controls using GSEA. P-values were calculated using the Benjamini and Hochberg False Discovery method. (c) Percent γH2AX-positive nuclei (top) and percent pRPA32-positive nuclei (bottom) per tumor area by IHC. Images are 40x with bar=100μm. P-values were calculated using unpaired t test with Welch’s correction. (d) Comparison of GSVA scores for the KEGG cytosolic DNA pathway transcripts between cases and controls. P-value was calculated using unpaired t test with Welch’s correction. (e) Expression of each gene in the KEGG cytosolic DNA Sensing Pathway with pro-inflammatory functions compared between case and control groups. P-value for each comparison was calculated using unpaired t test.
