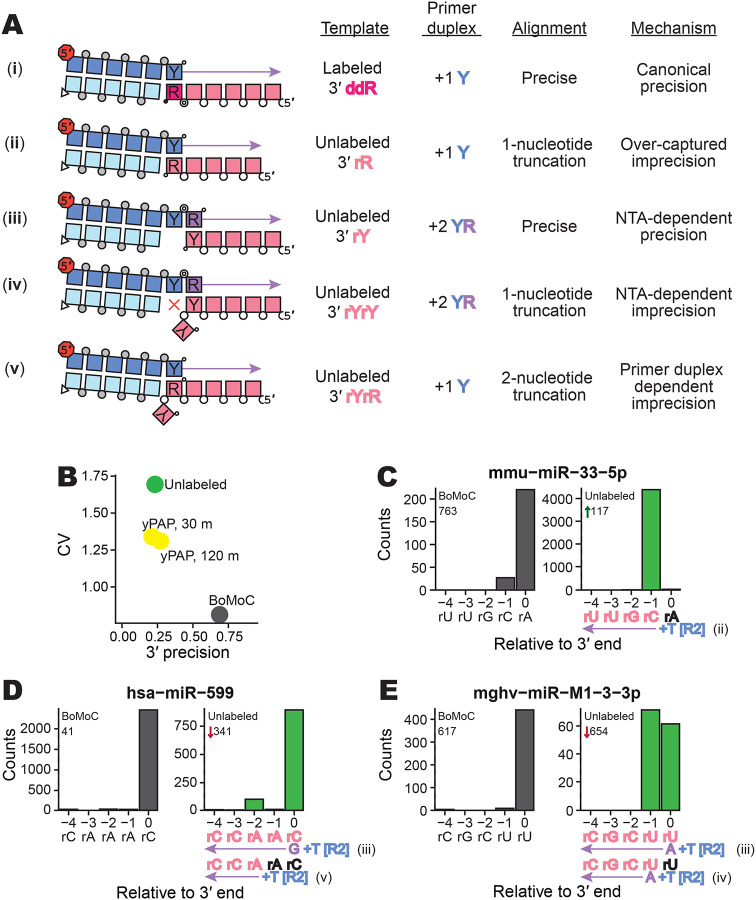
Figure 2:
Evaluating imprecision of end-to-end sequence capture at RNA 3′ ends. A, Mechanisms, numbered by Roman numeral, of 3′-end capture in OTTR. Column one illustrates five specificities for capturing an input-template 3′ end, following the color legend from Figure 1. Phosphodiester bonds are represented by circles: gray for chemically synthesized bonds, concentric double circle for BoMoC-polymerized, unfilled for input template RNA. Strand 3′ end symbols are a small black circle for dideoxynucleotide, a small open circle for 3′-OH, and a triangle for 3′-OH replacement with an unextendible carbon linker. The red octagon indicates the presence of a 5′ fluorescent dye. Column two specifies the labeling status of the input-template 3′ end. Column three identifies the primer-duplex 3′ overhang supporting a template jump. Column four details the impact on 3′ sequence coverage. Column five categorizes the 3′-end capture mechanism.
B, Library-wide CV of miRXplore miRNA counts and mean fraction of alignments with complete 3′-end coverage (3′ precision). BoMoC, yPAP (for 30 or 120 minutes), or no enzyme (Unlabeled) was used for 3′ labeling.
C-E, Panel pairs depict 3′-end coverage with BoMoC (left) or no enzyme (right) for 3′ labeling. The miRNA sequence and name, and relative rank-order, are indicated at top. At bottom, position “0” is the miRNA 3′ end. Right panels have at bottom the miRNA sequence color-coded by alignment inclusion (pink) or exclusion (black). The Read2 (R2) primer sequence and adjacent +1 T overhang (+T) are in blue, and reference to the capture mechanism detailed in (A) is indicated. Input RNA 3′ sequence excluded by adapter trimming is indicated in black. The miRNA mmu-miR-33–5p is miRBase: MIMAT0000667, the miRNA hsa-miR-599 is miRBase: MIMAT0003267, and the miRNA mghv-miR-M1–3-3p is miRBase: MIMAT0001566.