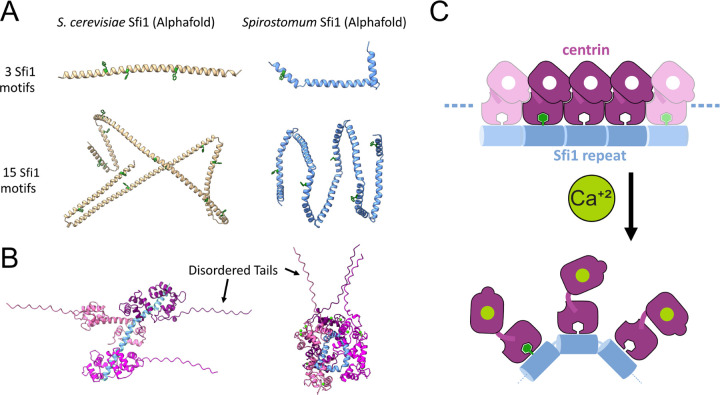
FIG. 5.
Protein structures predicted by AlphaFold suggest a molecular mechanism of Sfi1-centrin mediated contraction. A Comparison between AlphaFold-predicted structures of S. cerevisiae and Spirostomum Sfi1, each pair shown at approximately the same magnification. Tryptophan residues are highlighted in green. The model predicts a large number of bending sites throughout Spirostomum Sfi1 in comparison to S. cerevisiae Sfi1 (Supp. Data 1–4). B Examples of AlphaFold-predicted structures of Spirostomum Sfi1-centrin interactions that may regulate persistence length. Predictions show two predicted states, one with a stabilized Sfi1, similar to the S. cerevisiae crystal structure, and the other with kinked Sfi1, similar to the predicted structure in the absence of centrin (Supp. Data 5 and 6). C Proposed molecular mechanism for contraction of the centrin/Sfi1 complex. In the absence of calcium, centrin may stabilizes Sfi1 in an elongated conformation. With the addition of calcium, centrin-centrin and/or centrin-Sfi1 interactions change, allowing Sfi1 to bend and shortening its end-to-end distance.