Abstract
Background:
Early detection of cognitive decline during the preclinical stage of Alzheimer’s disease is crucial for timely intervention and treatment. Clinical notes, often found in unstructured electronic health records (EHRs), contain valuable information that can aid in the early identification of cognitive decline. In this study, we utilize advanced large clinical language models, fine-tuned on clinical notes, to improve the early detection of cognitive decline.
Methods:
We collected clinical notes from 2,166 patients spanning the 4 years preceding their initial mild cognitive impairment (MCI) diagnosis from the Enterprise Data Warehouse (EDW) of Mass General Brigham (MGB). To train the model, we developed SCD-Tron, a large clinical language model on 4,949 note sections labeled by experts. For evaluation, the trained model was applied to 1,996 independent note sections to assess its performance on real-world unstructured clinical data. Additionally, we used explainable AI techniques, specifically SHAP values, to interpret the models predictions and provide insight into the most influential features. Error analysis was also facilitated to further analyze the model’s prediction.
Results:
SCD-Tron significantly outperforms baseline models, achieving notable improvements in precision, recall, and AUC metrics for detecting Subjective Cognitive Decline (SCD). Tested on many real-world clinical notes, SCD-Tron demonstrated high sensitivity with only one false negative, crucial for clinical applications prioritizing early and accurate SCD detection. SHAP-based interpretability analysis highlighted key textual features contributing to model predictions, supporting transparency and clinician understanding.
Conclusion:
SCD-Tron offers a novel approach to early cognitive decline detection by applying large clinical language models to unstructured EHR data. Pretrained on real-world clinical notes, it accurately identifies early cognitive decline and integrates SHAP for interpretability, enhancing transparency in predictions.
Keywords: Cognitive decline, Alzheimer’s disease, large language model, clinical notes, electric health records, explainable AI
1. Introduction
Alzheimers disease (AD) is the most common form of dementia and ranks among the top 10 leading causes of death in the United States [1, 2]. It is projected that by 2060, nearly 14 million people will be affected by Alzheimer’s disease and related dementias (ADRD) [3, 4]. AD is a neurodegenerative disorder characterized by a gradual decline in memory and other cognitive functions. The disease typically progresses through three stages: preclinical, mild cognitive impairment (MCI), and dementia [5, 6]. Early detection of AD is crucial as it can significantly slow the progression of dementia symptoms, thereby improving the quality of life for patients and their caregivers [7, 8].
In recent decades, various methods utilizing different data modalities, such as MRI, PET scans, cerebrospinal fluid (CSF) analysis, and genetic testing, have been developed for detecting cognitive decline [9, 10, 11, 12]. However, these methods are often invasive, expensive, and resource-intensive, which can limit their feasibility for large-scale screening. In contrast, Electronic Health Records (EHRs) offer several advantages for cognitive decline detection [13, 14]. EHRs are non-invasive, cost-effective, and readily accessible, making them a more practical option for widespread screening compared to high-cost imaging and laboratory-based tests. Additionally, EHRs contain extensive unstructured clinical notes that can capture subtle cognitive symptoms that may be overlooked by other methods, enabling earlier detection and intervention. Thus we focus on using the unstructured EHR data, i.e., clinical notes for our study.
Natural Language Processing (NLP) has emerged as a powerful tool for extracting meaningful insights from unstructured data in EHRs, particularly from clinical notes [15, 16]. NLP facilitates the automatic analysis of large volumes of text, enabling the identification of patterns, relationships, and trends that are often difficult to detect manually [17, 18]. In recent years, transformer-based models like BERT [19] and its domain-specific variants such as ClinicalBERT [20], BioBERT [21] and MedBERT [22], has significantly enhanced the ability to extract relevant information for clinical decision-making [23, 24, 25]. Some prior studies have applied these techniques for AD or MCI-to-AD conversion prediction [26, 27, 28]. However, these models have several limitations in clinical applications. They are not trained on large-scale unstructured clinical notes, leading to an incomplete understanding of specialized medical terminology. Additionally, even with fine-tuning on small medical datasets, they lack the depth of clinical knowledge needed for complex healthcare tasks. These limitations highlight the need for more advanced clinical language models in our study.
To overcome these challenges, recent developments in large language models (LLMs) have achieved great breakthrough [29, 30, 31], and also introduced more sophisticated architectures tailored for the medical domain [32, 33, 34]. In a previous study in our group, we have already tried general-domain large language models (e.g., GPT and Llama) [35]. Despite this potential, there have been relatively few studies leveraging large clinical language models specifically designed for the early detection of cognitive decline in the preclinical stage of Alzheimer’s disease (AD) using clinical notes. Thus this work focuses on pre-trained models on medical records. Recently, models like GatorTron [36] and NYUTron [37] represent a significant leap forward, addressing the limitations of the above-mentioned models. These LLMs are trained on vastly larger and more diverse clinical datasets, encompassing a broad range of medical and general knowledge, which enhances their ability to handle biomedical related tasks. Our study aims to address this gap by demonstrating the potential of large clinical language models for efficient screening of AD at its earliest stages. This approach opens the door to more scalable and effective screening tools for brain health.
Based on the above motivations, we propose an AI model, SCD-Tron, built upon a large clinical language model for detecting cognitive decline at the preclinical stage of Alzheimer’s disease (AD) using unstructured EHR data, specifically clinical notes. Our approach leverages the power of a state-of-the-art clinical language model to capture the complex clinical knowledge and nuanced semantics within these notes. To further enhance the interpretability of the model, we incorporate explainable AI techniques using SHAP values, offering valuable insights into the factors influencing the models predictions.
This paper makes the following contributions.
Integration of a Large Clinical Language Model: Our work is one of the first to adapt large clinical language model, specifically for the early detection of cognitive decline using unstructured clinical notes. This represents a significant step forward in leveraging LLMs for preclinical stage of Alzheimer’s disease.
Explainability in Clinical Context: By applying SHAP to interpret the models predictions, we introduce a novel way to gain insights into how large clinical language models make decisions. This approach not only enhances transparency but also provides actionable information that clinicians can use to understand and trust the models outputs.
Real-World Data Application: Our study applies these advanced techniques to a real-world dataset of unstructured EHR notes, demonstrating the practical utility of our model in clinical settings.
2. Materials
2.1. Data Collection
This study was conducted using data from Mass General Brigham (MGB), the largest healthcare system serving the Greater Boston area. Clinical notes for patients were retrieved from MGB’s Enterprise Data Warehouse (EDW). We specifically extracted clinical notes from the four years preceding each patients initial mild cognitive impairment (MCI) diagnosis in 2019 to focus on the early stages of cognitive decline. The data usage has been approved by the Institutional Review Board (IRB) of MGB (Number: 2022P002987, 2022P002772), and all of them were deidentified before our model development.
2.2. Data Processing
The clinical notes were divided into sections using the NLP tool, i.e., Medical Text Extraction, Reasoning, and Mapping System (MTERMS) [38]. Given the variability in the length and content of clinical notes, these sections can range from highly relevant to irrelevant in terms of cognitive assessment. For annotation, sections strongly associated with progressive cognitive decline or those consistent with the onset of mild cognitive impairment (MCI) were labeled as positive. Sections deemed irrelevant, uncertain, or related to reversible conditions (e.g., cognitive improvements or impairments due to injury or surgery) were labeled as negative.
2.3. Training and Test Sets Setting
All the data (sections in clinical notes) were split into a training set (Data Set I) and a separate test set (Data Set II). The training set contained 4, 949 sections among which a total of 1, 453 were labeled as subjective cognitive decline. The test set consisted of 1, 996 separated sections from the training set, and there were 69 sections were labeled as subjective cognitive decline.
3. Methods
3.1. Overview
The overall pipeline of our framework is illustrated in Fig. 1. Our proposed framework, SCD-Tron, was designed to detect early cognitive decline using unstructured clinical notes. All the notes were firstly segmented into different sections which were fed into to the AI model. A large pre-trained clinical language model was leveraged to extract the sematic features of each section. Finally, a binary classification network output the prediction of cognitive decline status (SCD or non-SCD). To enhance the interpretability of the proposed model, an explainable AI technique, i.e., SHAP (SHapley Additive exPlanations) was integrated into the framework. The SHAP values can provide interpretability for the model’s predictions.
Figure 1:
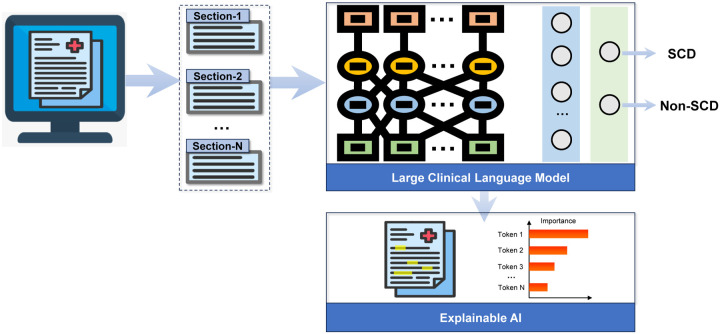
Illustration of the framework and workflow of the proposed SCD-Tron based on large clinical language model for subjective cognitive decline detection.
3.2. Model Structure
The overall structure of the proposed SCD-Tron model is shows in Fig. 1. The SCD-Tron model was built upon GatorTron, a state-of-the-art large clinical language model pre-trained on a vast corpus of clinical notes. The Transformer [39] served as the core structure of the model. The pre-training allowed the model to capture complex clinical knowledge and nuances present in the unstructured notes. We trained the model for the specific task of cognitive decline detection by feeding clinical note sections into the model and applying a classification head to output a binary prediction (SCD or non-SCD). To ensure interpretability, SHAP values were employed to explain model decisions, allowing the identification of the most influential tokens that contribute to each prediction.
3.3. Training
The SCD-Tron model was trained on a dataset of clinical notes collected from patients at Mass General Brigham (MGB). During training, the model processeed clinical note sections as input and learned to predict whether the patient has subjective cognitive decline (SCD) or not. During training, an initial learning rate of 2e-5 was used, controlled by the AdamW optimizer. This learning rate allowed for gradual adjustments to the model weights without drastic changes.
The model was trained on a standard supervised classification task with binary cross-entropy loss as the objective function. A linear learning rate scheduler with warm-up was employed to ensure smooth convergence by slowly increasing the learning rate at the beginning of training and gradually decreasing it later. We set the maximum input length of clinical note sections to 512 tokens, enabling the model to handle lengthy text inputs common in clinical notes. We found that reducing the input length can negatively impact performance. The model was trained for 2 epochs, with each epoch involving the complete pass of the training data. This was found to provide sufficient convergence without overfitting. The proposed framework was implemented by the PyTorch and HuggingFace Transformer library [40].
3.4. Inference and Explainability
Inference was performed by applying the trained SCD-Tron model to unseen clinical notes from the test set. For each note, the model predicted whether the patient was likely to have cognitive decline or not. To provide transparency in decision-making, we employed SHAP (SHapley Additive exPlanations) values [41, 42] to interpret the models outputs.
SHAP values allowed us to identify which sections or tokens in the clinical notes contributed most to the model’s decision. By assigning importance scores to each feature (word or token), SHAP offered a detailed breakdown of the factors influencing the prediction. This interpretability is particularly important in the clinical context, where understanding the rationale behind a decision is critical for clinician trust and model adoption. Visualizing these SHAP values enhanced the overall transparency of the SCD-Tron model, allowing clinicians to better understand why the model predicts SCD for a particular patient, fostering confidence in its predictions for real-world clinical applications.
In large language models (LLMs), another option for interpretation is using prompting to generate detailed explanations in sentence form. However, while prompts are commonly used by generative language models to create narrative explanations based on input text, they are not ideally suited for the structured interpretation required in classification tasks. Instead, we chose SHAP for its ability to precisely quantify the contribution of each input feature (word or token) to the model’s decision. SHAP values provide a clear, feature-level breakdown of the factors influencing the prediction, which is crucial in clinical contexts where understanding the specific reasons behind a decision is essential for clinician trust and model adoption. Additionally, prompt-based explanations are more limited in their application, particularly within LLMs, whereas SHAP is a model-agnostic tool. This means SHAP can be used to explain predictions from any machine learning model, significantly enhancing the flexibility and applicability of our approach.
4. Experiment
4.1. Experimental Setup
4.1.1. Evaluation Metrics
The task in our study was to identify if a clinical section was relevant to progressive cognitive decline or not. A section that represented cognitive decline was regarded as the positive case (with label 1), while the others were negative cases (with label 0). It was a binary classification problem. For performance evaluation, we used four metrics, i.e., Precision, Recall, F1-score, Receiver Operating Characteristic Area Under the Curve (ROC-AUC), and Area under the Precision-Recall Curve (PR-AUC). Higher values for each evaluation metric indicated better performance.
4.1.2. Competing Methods
Logistic Regression:
We represented each section using Term Frequency-Inverse Document Frequency (TF-IDF) based on n-grams (n=1, 2). These feature vectors were then utilized to train a logistic regression classifier, which predicted whether a section indicates cognitive decline.
Random Forest:
We used the TF-IDF features of the sections of clinical notes to train a Random Forest classifier for the task of cognitive decline detection. The key hyperparameter, i.e., number of trees in the forest, was set to 100.
Support Vector Machine (SVM):
TF-IDF features of the sections were used to train an SVM classifier (with a linear kernel) for cognitive decline detection.
BERT:
We used the vanilla BERT for the SCD detection task. The BERT model was first fine-tuned on the training set, and then applied to the test set. A fully-connected layer was used as the classification head for SCD vs. non-SCD classification on the sections.
BioBERT:
The BioBERT model was pre-trained on biomedical literature (e.g., scientific papers from PubMed). We fine-tuned it on the training set, and then applied to the test set. A fully-connected layer was added as the classifier for SCD vs. non-SCD classification on the sections.
ClinicalBERT:
The ClinicalBERT was pre-trained on the MIMIC III dataset which contained clinical notes, including discharge summaries, progress notes, etc. A fully-connected layer was built for the identification of SCD on the sections.
In our experiments with these deep learning-based methods, each model was run five times, and the average and standard deviation of the results were reported.
4.2. Result
The cognitive decline detection results of the proposed SCD-Tron model and several other competing methods are shown in Table 1.
Table 1:
Performance of Various Methods in the Task of Cognitive Decline Detection
| Method | Precision | Recall | F1-Score | AUC-ROC | AUC-PR |
|---|---|---|---|---|---|
| Logistic Regression | 0.8163 | 0.5797 | 0.6780 | 0.9647 | 0.7383 |
| Random Forest | 0.8143 | 0.8261 | 0.8201 | 0.9955 | 0.8936 |
| SVM | 0.7419 | 0.6667 | 0.7023 | 0.9670 | 0.7419 |
| BERT | 0.8114±0.0245 | 0.9652±0.0116 | 0.8811±0.0118 | 0.9967±0.0003 | 0.9338±0.0114 |
| BioBERT | 0.8399±0.0336 | 0.9449±0.0169 | 0.8886±0.0137 | 0.9983±0.0004 | 0.9614±0.0042 |
| ClinicalBERT | 0.8114±0.0265 | 0.9536±0.0108 | 0.8764±0.0136 | 0.9981±0.0004 | 0.9517±0.0093 |
| SCD-Tron (Ours) | 0.8812±0.0244 | 0.9826±0.0108 | 0.9289±0.0138 | 0.9993±0.0002 | 0.9804±0.0074 |
We also plot the ROC curves and Precision-Recall curves of our SCD-Tron and other models in the task of SCD detection, as shown in Fig. 2.
Figure 2:
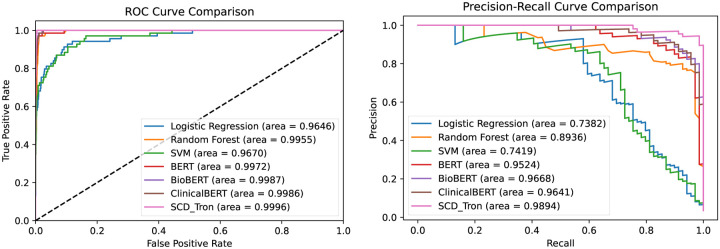
Comparison of ROC curve and precision-recall curve of different methods in the task of SCD identification.
From the experimental results, we have the following observations.
Compared with the conventional features engineering (i.e., TF-IDF features), the pre-trained language models achieved significantly higher performance. This demonstrated the superior feature learning capabilities of deep learning-based models, particularly in capturing complex patterns from unstructured clinical notes data.
The pre-trained language models achieved notably higher recall compared to other methods, demonstrating their ability to effectively identify progressive SCD cases. This is particularly valuable in clinical applications, where it is crucial to prioritize the comprehensive detection of patients at risk for Alzheimer’s disease, even at the expense of some false positives.
The proposed SCD-Tron model outperformed other pre-trained language models across all evaluation metrics. This can be attributed to SCD-Tron’s pre-training on a significantly larger corpus of clinical notes compared to the other models, which were trained on plain texts, medical articles, or smaller datasets of clinical notes. These results underscored the advantage of large-scale clinical language models in effectively processing unstructured EHR data.
5. Discussion
5.1. Explaining Model Predictions Using SHAP Values
To better understand the decision-making process of the proposed SCD-Tron model, we utilized SHAP (SHapley Additive exPlanations) to interpret its predictions. SHAP values provided a visual breakdown of how each token or word in the clinical notes contributes to the models decision, helping to explain the predictions for progressive SCD cases.
As shown in Fig. 3, we visualized the SHAP values and highlighted the relevant text for two individual sections that were correctly classified as positive samples (i.e., SCD) by SCD-Tron. The result revealed that certain texts related to memory loss strongly influenced the model’s prediction, demonstrating how the model identifies key clinical features in the notes that were indicative of cognitive decline.
Figure 3:
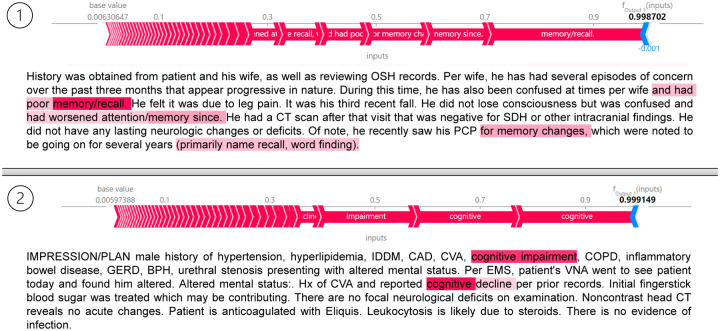
SHAP values with highlighted tokens in two individual sections that have been correctly classified as positive samples (i.e., SCD) by the proposed SCD-Tron model.
5.2. Key Words that Influence Model Prediction
In addition to analyzing individual predictions, SHAP values can be used to identify common key words across the entire test set that significantly influence the models predictions for progressive SCD cases. By aggregating SHAP values for all identified positive samples, we can generate an overall view of which terms contribute most strongly to the model’s decision-making process.
For this analysis, we computed the SHAP values for all positive samples in the test set and average them to assess the overall contribution of different tokens. This process helped highlight the most influential words that consistently appeared in the clinical notes related to SCD predictions. As visualized in Fig. 4, terms related to memory, cognition, and behavioral changes (e.g., fluency) emerged as key factors driving the models predictions. This aggregated explanation provided clinicians with valuable insight into the underlying patterns the model detects in unstructured clinical notes.
Figure 4:
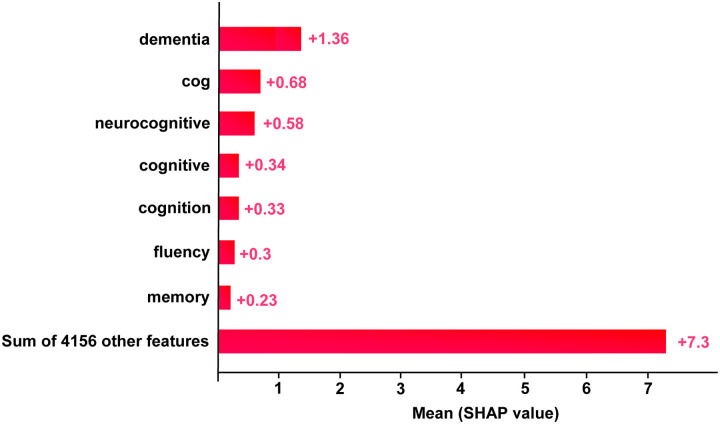
Importance features for SCD identification in terms of SHAP values selected by the proposed SCD-Tron model.
5.3. Error Analysis
To further understand the prediction of the proposed SCD-Tron model, we conducted error analysis by exploring the false positive and false negative prediction cases by the model.
We first visualized the confusion matrix of the model’s prediction result, as shown in Fig. 5. From the results, it is evident that the model achieves high sensitivity, with only one false negative case. This is crucial in clinical practice, as missing positive cases can have more severe consequences than false alarms. On the other hand, there are 8 false positive predictions.
Figure 5:
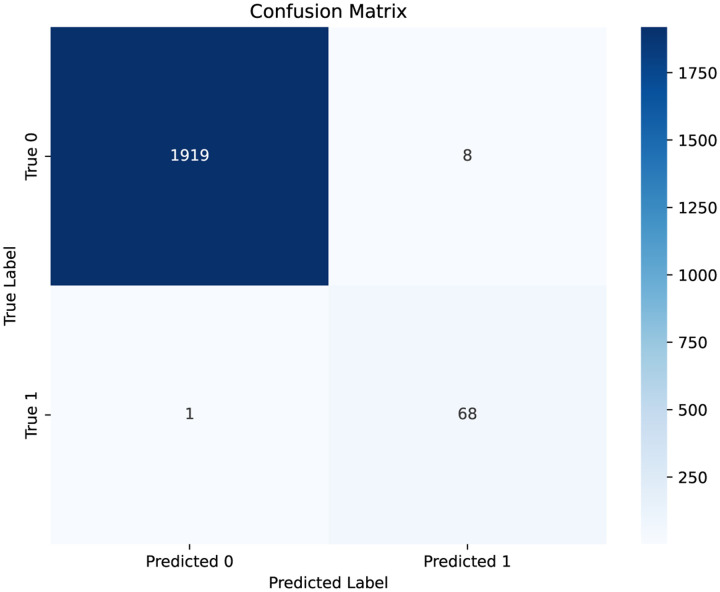
Confusion matrix of the model’s prediction. Label 1 represents SCD while label 0 denotes non-SCD.
Table 2 presents the representative note sections corresponding to both the false negative and false positive cases, along with explanations for the errors made by the model.
Table 2:
Error Analysis for False Negative/Positive Predictions of the SCD-Tron Model
| Potential Reasons | Representative Cases |
|---|---|
| Weak Evidence. Primarily focuses on physical impairments and care goals, with minimal mention of cognitive symptoms, potentially leading to misclassification |
False Negative. GOALS HH - ADL / IADL Impairment. HH - Promote higher level of independence with performance of ADLs/IADLs. [OT]. HH - Advance Care Planning. HH - Verbalize awareness of Advance Care Planning. [MSW, OT, PT, SLP, SN]. HH - Aspiration - Risk of. HH - Knowledge and management of aspiration precautions. [MSW, OT, PT, SLP, SN]. … HH - Health Maintenance / Patient Strengths Impacting Goal Achievement. HH - Patient will utilize their strengths to achieve optimal knowledge and management for safe home/ community functioning. [MSW, OT, PT, SLP, SN]. HH - Mental Status - Impaired. HH - Mental status allows patient to safely function within their home environment… |
| The issue is caused by side effects of medication rather than SCD. |
False Positive. 1 Onc:. Clinically doing well on Ibrutinib. However S and her husband are concerned that her memory and anxiety/depression has worsened on this medication. We discussed this with Dr. A and Dr. B as well. We offered holding the ibrutinib to see if this would help. However she is not interested in doing this at this time. We discussed other options such as Venetoclax. She prefers to treat the depression instead and stay on Ibrutinib. Please see Dr. A’s note from today regarding plan for f/u… |
5.4. Limitations
While SCD-Tron demonstrated promising results in detecting early cognitive decline, several limitations should be acknowledged. First, our study was based on a specific dataset from a single healthcare system, which may limit the model’s applicability to other populations from other healthcare systems. Furthermore, SCD-Tron primarily focused on text data, which limits its ability to integrate other valuable data modalities, such as imaging or biomarkers, that could provide a more comprehensive assessment of cognitive decline.
Future work will focus on expanding the scope of SCD-Tron to incorporate multi-modal data, such as MRI scans or genetic information, to improve detection accuracy. Additionally, we plan to extend the model’s validation to broader and more diverse patient populations across different healthcare systems to ensure its robustness and generalizability.
6. Conclusion
In this paper, we present SCD-Tron, a novel approach for the early detection of cognitive decline using unstructured clinical notes from real-world electronic health records (EHR). By leveraging the power of large-scale clinical language models, SCD-Tron is designed to detect early signs of Subjective Cognitive Decline (SCD), a critical precursor to Alzheimer’s disease. Our approach adapts state-of-the-art clinical language models to handle the complexities of unstructured clinical data, providing a robust tool for early diagnosis and intervention. Additionally, the integration of SHAP for model interpretability enhances the transparency of predictions, offering clinicians valuable insights into the factors influencing the models decisions. This combination of advanced modeling and explainability makes SCD-Tron a trustworthy and impactful tool for clinical practice.
Acknowledgments
This study was funded by NIH-NIA R44AG081006 and R01AG080429.
References
- [1].Masters C. L., Bateman R., Blennow K., Rowe C. C., Sperling R. A., Cummings J. L., Alzheimer’s disease, Nature Reviews Disease Primers 1 (1) (2015) 1–18. [Google Scholar]
- [2].Ahmad F. B., Anderson R. N., The leading causes of death in the US for 2020, JAMA 325 (18) (2021) 1829–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kramarow E. A., Tejada-Vera B., National vital statistics reports, National Vital Statistics Reports 68 (2) (2019) 1–18. [Google Scholar]
- [4].Nandi A., Counts N., Bröker J., Malik S., Chen S., Han R., Klusty J., Seligman B., Tortorice D., Vigo D., et al. , Cost of care for Alzheimers disease and related dementias in the United States: 2016 to 2060, NPJ Aging 10 (1) (2024) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Scharre D. W., Preclinical, prodromal, and dementia stages of Alzheimer’s disease, Practical Neurology 15 (2019) 36–47. [Google Scholar]
- [6].Rabin L. A., Smart C. M., Amariglio R. E., Subjective cognitive decline in preclinical Alzheimer’s disease, Annual review of clinical psychology 13 (1) (2017) 369–396. [Google Scholar]
- [7].Crous-Bou M., Minguillón C., Gramunt N., Molinuevo J. L., Alzheimers disease prevention: from risk factors to early intervention, Alzheimer’s research & therapy 9 (2017) 1–9. [Google Scholar]
- [8].Robinson L., Tang E., Taylor J.-P., Dementia: timely diagnosis and early intervention, BMJ 350 (2015). [Google Scholar]
- [9].Arrondo P., Elía-Zudaire Ó., Martí-Andrés G., Fernández-Seara M. A., Riverol M., Grey matter changeson brain MRI in subjective cognitive decline: A systematic review, Alzheimer’s Research & Therapy 14 (1) (2022) 1–16. [Google Scholar]
- [10].Lagarde J., Olivieri P., Tonietto M., Tissot C., Rivals I., Gervais P., Caillé F., Moussion M., Bottlaender M., Sarazin M., Tau-PET imaging predicts cognitive decline and brain atrophy progression in early Alzheimer’s disease, Journal of Neurology, Neurosurgery & Psychiatry 93 (5) (2022) 459–467. [DOI] [PubMed] [Google Scholar]
- [11].Wolfsgruber S., Polcher A., Koppara A., Kleineidam L., Frölich L., Peters O., Hüll M., Rüther E., Wiltfang J., Maier W., et al. , Cerebrospinal fluid biomarkers and clinical progression in patients with subjective cognitive decline and mild cognitive impairment, Journal of Alzheimer’s Disease 58 (3) (2017) 939–950. [Google Scholar]
- [12].Sherva R., Gross A., Mukherjee S., Koesterer R., Amouyel P., Bellenguez C., Dufouil C., Bennett D. A., Chibnik L., Cruchaga C., et al. , Genome-wide association study of rate of cognitive decline in Alzheimer’s disease patients identifies novel genes and pathways, Alzheimer’s & Dementia 16 (8) (2020) 1134–1145. [Google Scholar]
- [13].Li Q., Yang X., Xu J., Guo Y., He X., Hu H., Lyu T., Marra D., Miller A., Smith G., et al. , Early prediction of Alzheimer’s disease and related dementias using real-world electronic health records, Alzheimer’s & Dementia 19 (8) (2023) 3506–3518. [Google Scholar]
- [14].Yadav P., Steinbach M., Kumar V., Simon G., Mining electronic health records (EHRs): A survey, ACM Computing Surveys (CSUR) 50 (6) (2018) 1–40. [Google Scholar]
- [15].Hossain E., Rana R., Higgins N., Soar J., Barua P. D., Pisani A. R., Turner K., Natural language processing in electronic health records in relation to healthcare decision-making: A systematic review, Computers in Biology and Medicine 155 (2023) 1–24. [Google Scholar]
- [16].Wang L., Laurentiev J., Yang J., Lo Y.-C., Amariglio R. E., Blacker D., Sperling R. A., Marshall G. A., Zhou L., Development and validation of a deep learning model for earlier detection of cognitive decline from clinical notes in electronic health records, JAMA Network Open 4 (11) (2021) 1–11. [Google Scholar]
- [17].Khurana D., Koli A., Khatter K., Singh S., Natural language processing: state of the art, current trends and challenges, Multimedia Tools and Applications 82 (3) (2023) 3713–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li I., Pan J., Goldwasser J., Verma N., Wong W. P., Nuzumlalı M. Y., Rosand B., Li Y., Zhang M., Chang D., et al. , Neural natural language processing for unstructured data in electronic health records: a review, Computer Science Review 46 (2022) 1–29. [Google Scholar]
- [19].Devlin J., Chang M.-W., Lee K., Toutanova K., BERT: Pre-training of deep bidirectional transformers for language understanding, in: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies), Association for Computational Linguistics, 2019, pp. 4171–4186. [Google Scholar]
- [20].Huang K., Altosaar J., Ranganath R., ClinicalBERT: Modeling clinical notes and predicting hospital readmission, arXiv preprint arXiv:1904.05342 (2019). [Google Scholar]
- [21].Lee J., Yoon W., Kim S., Kim D., Kim S., So C. H., Kang J., BioBERT: a pre-trained biomedical language representation model for biomedical text mining, Bioinformatics 36 (4) (2020) 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rasmy L., Xiang Y., Xie Z., Tao C., Zhi D., Med-BERT: pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction, NPJ Digital Medicine 4 (1) (2021) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang B., Xie Q., Pei J., Chen Z., Tiwari P., Li Z., Fu J., Pre-trained language models in biomedical domain: A systematic survey, ACM Computing Surveys 56 (3) (2023) 1–52. [Google Scholar]
- [24].Hager P., Jungmann F., Holland R., Bhagat K., Hubrecht I., Knauer M., Vielhauer J., Makowski M., Braren R., Kaissis G., et al. , Evaluation and mitigation of the limitations of large language models in clinical decision-making, Nature Medicine (2024) 1–10. [Google Scholar]
- [25].Kalyan K. S., Rajasekharan A., Sangeetha S., Ammu: a survey of transformer-based biomedical pretrained language models, Journal of Biomedical Informatics 126 (2022) 1–23. [Google Scholar]
- [26].Mao C., Xu J., Rasmussen L., Li Y., Adekkanattu P., Pacheco J., Bonakdarpour B., Vassar R., Shen L., Jiang G., et al. , AD-BERT: Using pre-trained language model to predict the progression from mild cognitive impairment to Alzheimer’s disease, Journal of Biomedical Informatics 144 (2023) 1–8. [Google Scholar]
- [27].Oh I. Y., Schindler S. E., Ghoshal N., Lai A. M., Payne P. R., Gupta A., Extraction of clinical phenotypes for Alzheimer’s disease dementia from clinical notes using natural language processing, JAMIA Open 6 (1) (2023) 1–9. [Google Scholar]
- [28].Maclagan L. C., Abdalla M., Harris D. A., Stukel T. A., Chen B., Candido E., Swartz R. H., Iaboni A., Jaakkimainen R. L., Bronskill S. E., Can patients with dementia be identified in primary care electronic medical records using natural language processing?, Journal of Healthcare Informatics Research 7 (1) (2023) 42–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhao W. X., Zhou K., Li J., Tang T., Wang X., Hou Y., Min Y., Zhang B., Zhang J., Dong Z., et al. , A survey of large language models, arXiv preprint arXiv:2303.18223 (2023). [Google Scholar]
- [30].Minaee S., Mikolov T., Nikzad N., Chenaghlu M., Socher R., Amatriain X., Gao J., Large language models: A survey, arXiv preprint arXiv:2402.06196 (2024). [Google Scholar]
- [31].Naveed H., Khan A. U., Qiu S., Saqib M., Anwar S., Usman M., Akhtar N., Barnes N., Mian A., A comprehensive overview of large language models, arXiv preprint arXiv:2307.06435 (2023). [Google Scholar]
- [32].Thirunavukarasu A. J., Ting D. S. J., Elangovan K., Gutierrez L., Tan T. F., Ting D. S. W., Large language models in medicine, Nature Medicine 29 (8) (2023) 1930–1940. [Google Scholar]
- [33].Singhal K., Azizi S., Tu T., Mahdavi S. S., Wei J., Chung H. W., Scales N., Tanwani A., Cole-Lewis H., Pfohl S., et al. , Large language models encode clinical knowledge, Nature 620 (7972) (2023) 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nazi Z. A., Peng W., Large language models in healthcare and medical domain: A review, in: Informatics, Vol. 11, MDPI, 2024, p. 57. [Google Scholar]
- [35].Du X., Novoa-Laurentiev J., Plasaek J. M., Chuang Y.-W., Wang L., Marshall G., Mueller S. K., Chang F., Datta S., Paek H., et al. , Enhancing early detection of cognitive decline in the elderly: A comparative study utilizing large language models in clinical notes, medRxiv (2024). [Google Scholar]
- [36].Yang X., Chen A., PourNejatian N., Shin H. C., Smith K. E., Parisien C., Compas C., Martin C., Costa A. B., Flores M. G., et al. , A large language model for electronic health records, NPJ digital medicine 5 (1) (2022) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jiang L. Y., Liu X. C., Nejatian N. P., Nasir-Moin M., Wang D., Abidin A., Eaton K., Riina H. A., Laufer I., Punjabi P., et al. , Health system-scale language models are all-purpose prediction engines, Nature 619 (7969) (2023) 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhou L., Plasek J. M., Mahoney L. M., Karipineni N., Chang F., Yan X., Chang F., Dimaggio D., Goldman D. S., Rocha R. A., Using medical text extraction, reasoning and mapping system (MTERMS) to process medication information in outpatient clinical notes, in: AMIA Annual Symposium Proceedings, Vol. 2011, American Medical Informatics Association, 2011, pp. 1639–1648. [PMC free article] [PubMed] [Google Scholar]
- [39].Vaswani A., et al. , Attention is all you need, Advances in Neural Information Processing Systems (2017) 1–11. [Google Scholar]
- [40].Jain S. M., Hugging face, in: Introduction to transformers for NLP: With the hugging face library and models to solve problems, Springer, 2022, pp. 51–67. [Google Scholar]
- [41].Scott M., Su-In L., et al. , A unified approach to interpreting model predictions, Advances in Neural Information Processing Systems 30 (2017) 4765–4774. [Google Scholar]
- [42].Dwivedi R., Dave D., Naik H., Singhal S., Omer R., Patel P., Qian B., Wen Z., Shah T., Morgan G., et al. , Explainable AI (XAI): Core ideas, techniques, and solutions, ACM Computing Surveys 55 (9) (2023) 1–33. [Google Scholar]


