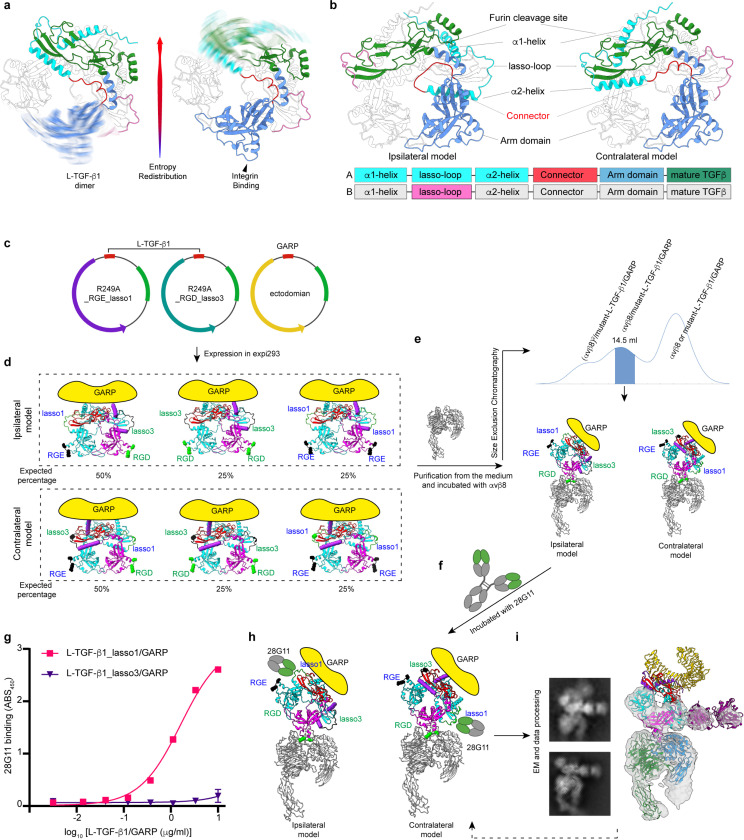
Figure 1.
Experiment design and determination of domain architecture of L-TGF-b1/GARP a, Ribbon diagram illustrating redistribution of conformational entropy in L-TGF-b1 from the RGD site to contralateral lasso loop prior (left) and after (right) avb8 binding7. b, Ribbon diagram illustrating two possible architectures of L-TGF-b1 dimer, in which the straitjacket domain of each prodomain is connected ipsilaterally (left) or contralaterally (right) with the arm domain. Color scheme in ribbon diagram and domain arrangements are the same. c, Plasmid constructs of L-TGF-b1_R249A_RGE_lasso1 (left), L-TGF-b1_R249A_RGD_lasso3 (middle) and GARP (right). d, Anticipated expression products and their proportions from the 1:1:1 transfection of Expi293 cells. Ribbon diagrams of L-TGF-b1 in ipsilateral (upper) or contralateral (bottom) architectures are predicted by AlphaFold. e, SEC profile (middle) of purified mutant L-TGF-b1/GARP incubated with avb8 ectodomain (left). The shaded peak contains two possible complexes (bottom). f, Incubation of monoclonal antibody clone 28G11 with the SEC purified complex. g, ELISA confirms that 28G11 only binds lasso1 but not lasso3. Shown is a single experiment with error bars showing SD from 2 experimental replicates. h, Two models illustrating binding of 28G11 to mutant L-TGF-b1/GARP in ipsilateral (left) and contralateral (right) architectures. i, Representative 2D class averages and 3D reconstruction with atomic models of a generic Fab and contralateral L-TGF-b1/GARP docked.