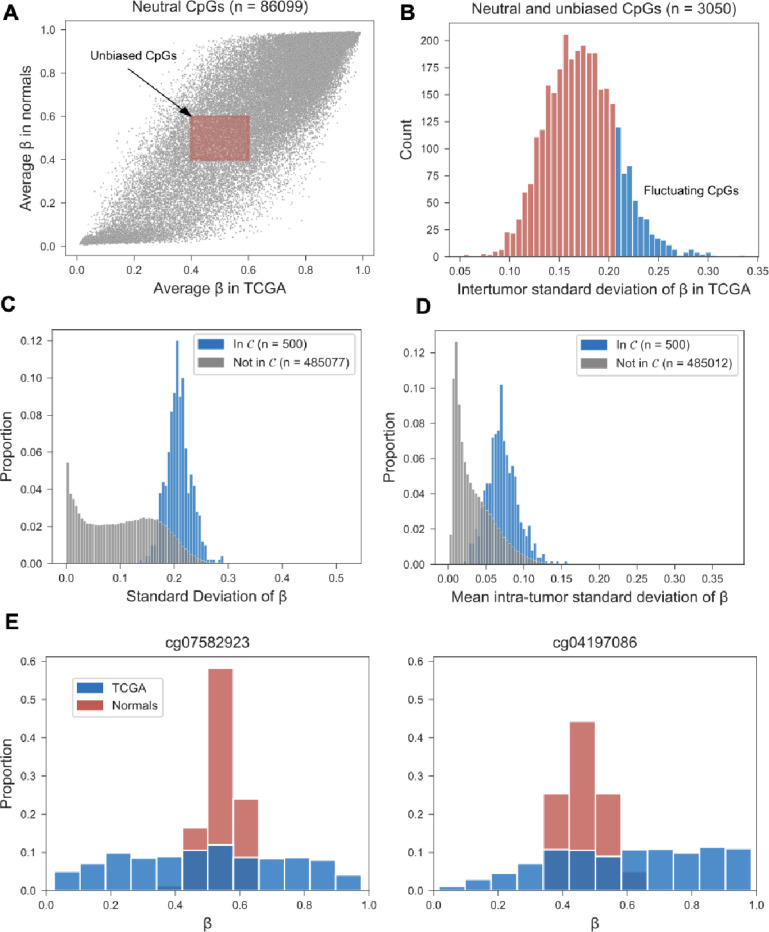
Figure 2. Selection and validation of unbiased fluctuating CpG (fCpG) sites.
(A) Starting with CpG sites that are nonregulatory and non-genic (gray dots), sites with an average methylation value between 0.4 and 0.6 in both breast cancers (N=634, TCGA cohort) and normal samples (N=79, Normal cohort) were considered unbiased (red shaded rectangle). (B) Sites were ranked by their inter-tumor standard deviation, and the top 500 were included the clock set . (C) In a separate cohort of breast cancers (N=146, Lund cohort), the inter-tumor standard deviation of was higher for fCp G sites in the clock set (median: 0.206) compared to CpGs not included in (median: 0.102; P=1×10−238, Wilcoxon rank-sum test). (D) In a small cohort of patients (N=5) with multiple primary samples (3–5 per patient), the intra-tumor standard deviation of was higher for CpG sites in the clock set (median: 0.070), compared to CpGs not included in (median: 0.026; P=6 0×10−179, Wilcoxon rank-sum test). (E) Distribution of the -values of two fCpG sites in the clock set , across breast cancers (N=634, TCGA cohort) and normal breast samples (N=76, Normal cohort).