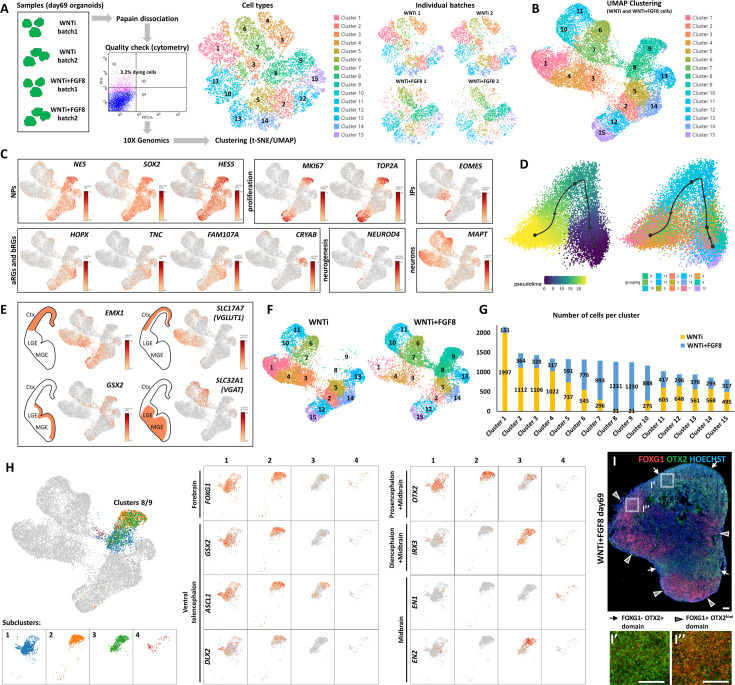
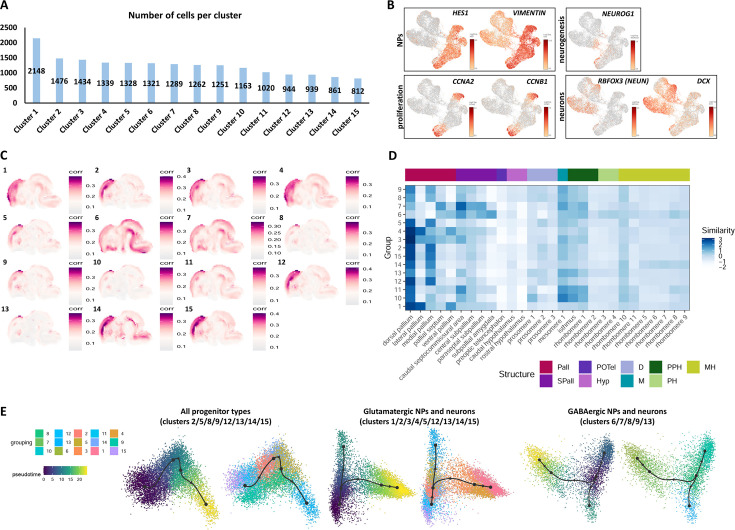
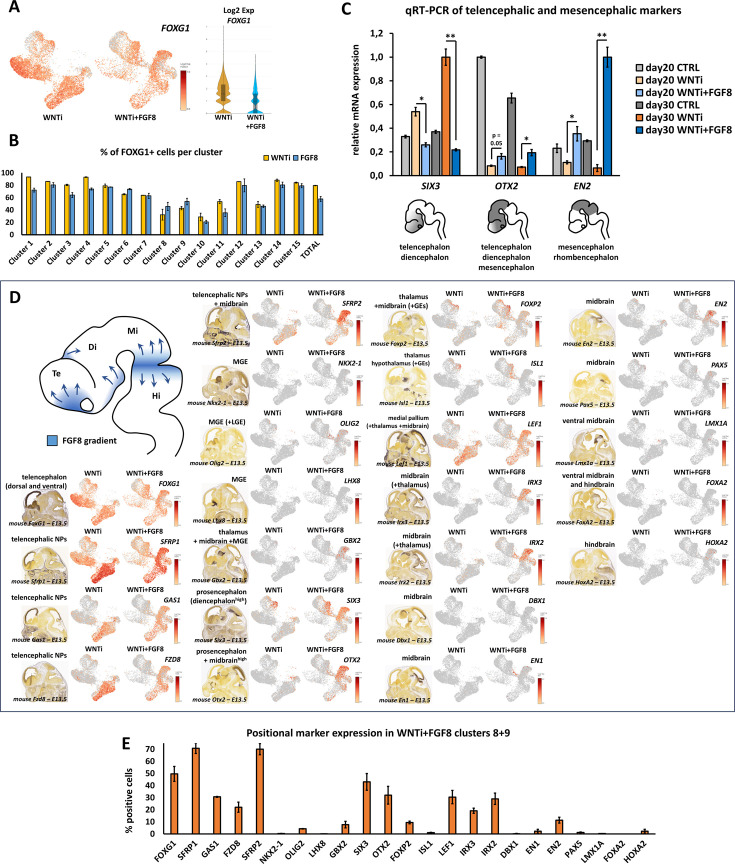
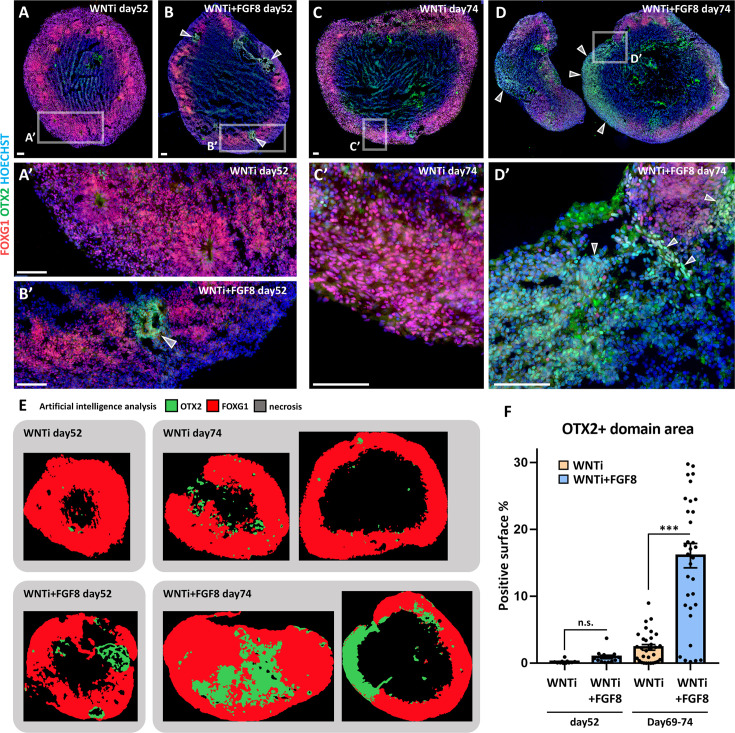
Figure 3. Single-cell RNA sequencing (scRNAseq) analysis of FGF8-induced cellular and molecular changes in human organoids.
(A) Experimental setup for scRNA-seq analysis of control (WNTi) and FGF8-treated (WNTi + FGF8) telencephalic organoids at day 69. Two independent batches of WNTi and WNTi + FGF8 organoids (each containing 2–3 organoids) were dissociated into single cells and processed using Chromium (10 X Genomics technology). Cells were clustered and visualized in 2D space using t-SNE and UMAP algorithms. (B) UMAP clustering of WNTi and WNTi + FGF8 cells, identifying 15 distinct clusters. (C) Expression levels of known markers for different cell types, including neural progenitor cells (NPs: NESTIN, SOX2, HES5), proliferating progenitors (KI67, TOP2A), intermediate progenitors (IPs: EOMES), apical and basal radial glia cells (aRGs and bRGs: HOPX, TNC, FAM107A, CRYAB), and differentiating/differentiated neurons (NEUROD4 and MAPT, respectively). (D) Trajectory analysis showing the most probable developmental progression from NP clusters (2, 5, 8, 9, 12, 15) to post-mitotic cell types (notably clusters 1, 3, 4, 6, 7). (E) Expression level and cluster distribution of dorsal glutamatergic markers EMX1 and SLC17A7 (also called VGLUT1) and ventral GABAergic markers GSX2 and SLC32A1 (also called VGAT), indicating the coexistence of both glutamatergic and GABAergic NPs and neurons within FOXG1+ telencephalic organoids. (F, G) UMAP clustering of WNTi and WNTi + FGF8 cells shown separately, illustrating 15 distinct clusters and their respective proportions in each condition. Panel (G) shows the number of cells in each cluster originating from WNTi (yellow) or WNTi + FGF8 (blue) organoids. (H) UMAP projection of day69 organoid scRNA-seq data, identifying four cellular groups through sub-clustering analysis on WNTi + FGF8 clusters 8 and 9. Center and right panels display expression levels of markers for the forebrain (FOXG1), ventral telencephalon (GSX2, ASCL1 and DLX2), forebrain/midbrain (OTX2), diencephalon/mesencephalon (IRX3), and mesencephalon (EN1, EN2) across the four sub-clusters. (I-I’’) Immunostaining for FOXG1 (red) and OTX2 (green) in day69 WNTi + FGF8 organoids, showing distinct FOXG1+ and FOXG1- regions. White arrows indicate FOXG1- OTX2+ non-telencephalic areas (high magnification in I’), while arrowheads denote FOXG1+ OTX2low telencephalic areas (high magnification in I’’). Ctx, cortex; MGE, medial ganglionic eminence; LGE, lateral ganglionic eminence.