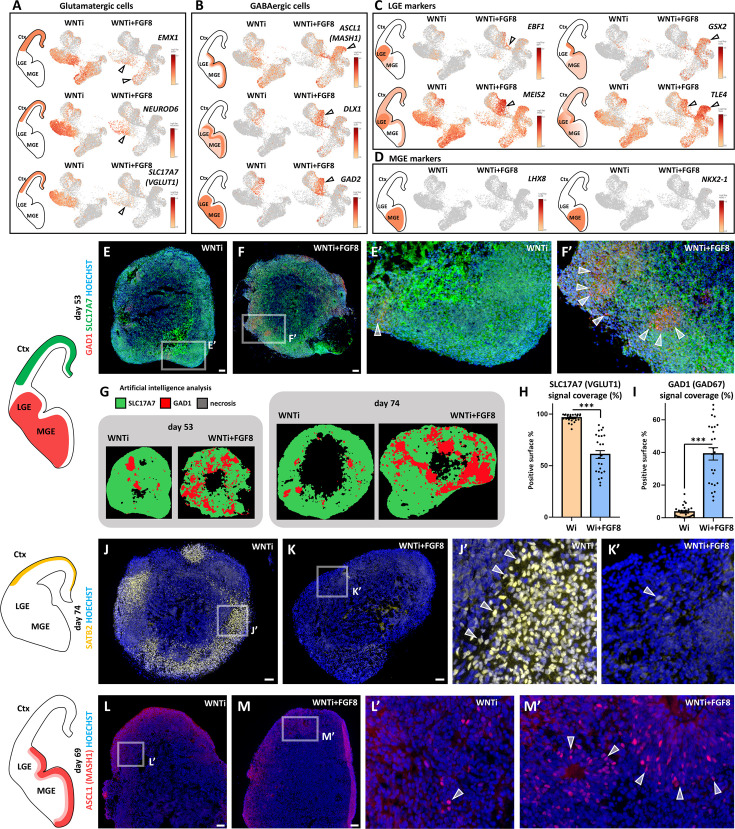
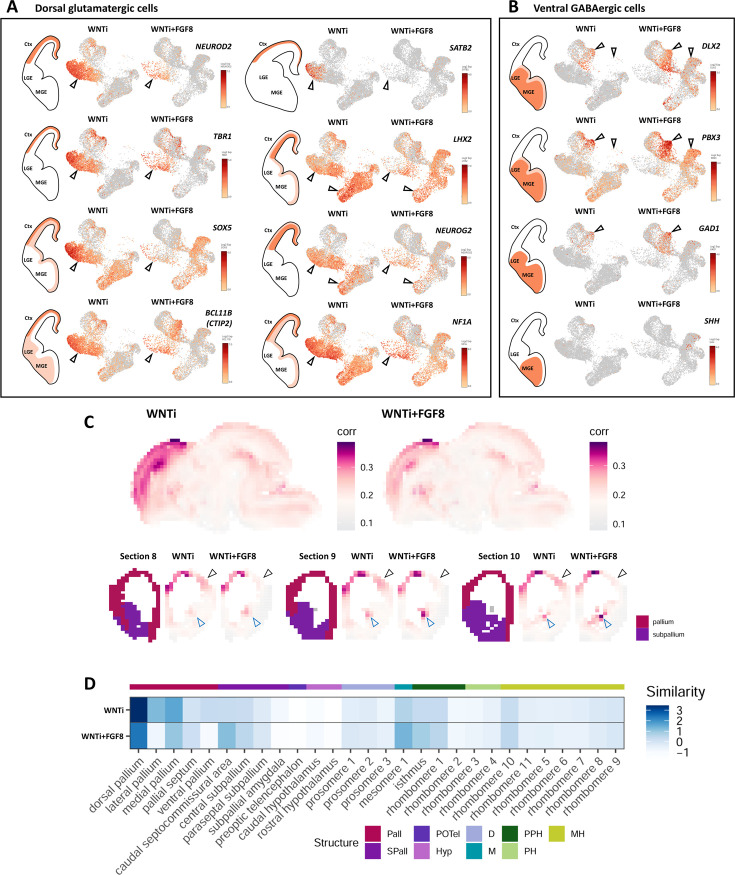
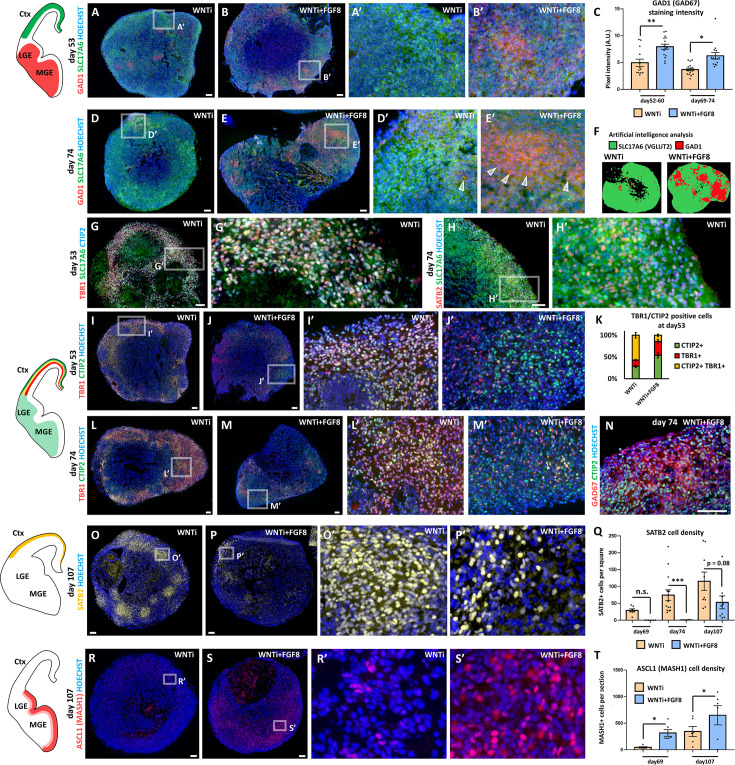
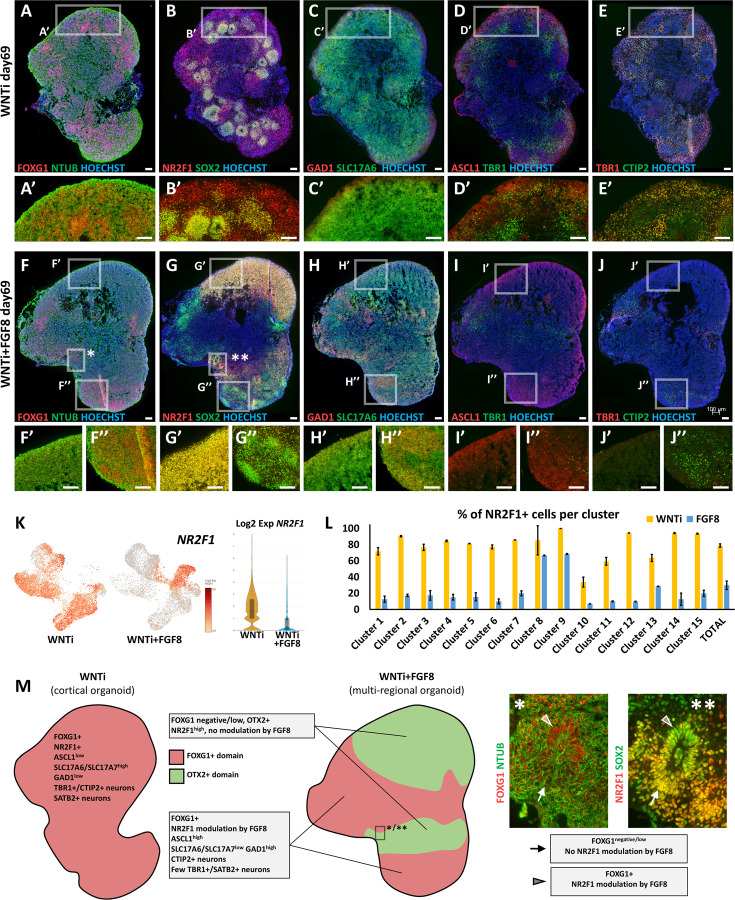
Figure 4. Effects of FGF8 treatment on the dorso-ventral cellular composition of telencephalic organoids.
(A, B) Expression levels of markers identifying dorsal glutamatergic NPs and neurons (Panel A; EMX1, NEUROD6, SLC17A7) and ventral GABAergic NPs and neurons (Panel B; ASCL1, DLX1, GAD2). Black arrowheads highlight clusters with the most notable changes in marker and cell abundance after FGF8 treatment. (C,D) Expression levels of known markers identifying ventral GABAergic NPs and neurons in the lateral ganglionic eminence (LGE) (Panel C; EBF1, GSX2, MEIS2 and TLE4) and the medial ganglionic eminence (MGE) (Panel D; NKX2-1 and LHX8). Black arrowheads in C highlight clusters with the largest differences in LGE marker and cell abundance following FGF8 treatment. (E-F’) GAD1 (red) and SLC17A7 (green) immunostaining in day53 control (WNTi) and treated (WNTi + FGF8) organoids, as indicated. The distribution of these markers in vivo is shown in the brain scheme on the left. (G–I) HALO software artificial intelligence (AI) analysis of marker distribution in day53 and day74 organoids. Representative images display areas automatically identified as SLC17A7+ (green), GAD1+ (red), and necrotic (black). Graphs in H and I show the proportions of SLC17A7+ (H) and GAD1+ (I) surface areas in day 74 organoid sections, as quantified by HALO AI; n≥8 sections from n≥4 organoids from n=1 batch per time point. (J-K’) Immunostaining for SATB2 (yellow) in day74 WNTi and WNTi + FGF8 organoids, as indicated. The left schematic depicts the in vivo distribution of SATB2+ neurons, and Figure 4—figure supplement 2 quantifies SATB2+ neuron density in these and additional samples. (L-M’) Immunostaining for ASCL1 (red) in day69 WNTi and WNTi + FGF8 organoids, as indicated. Left schematic shows the in vivo distribution of ASCL1+ ventral progenitors, with cell density detailed in Figure 4—figure supplement 2. Scale bars: 100 µm. Ctx, cortex; MGE, medial ganglionic eminence; LGE, lateral ganglionic eminence.