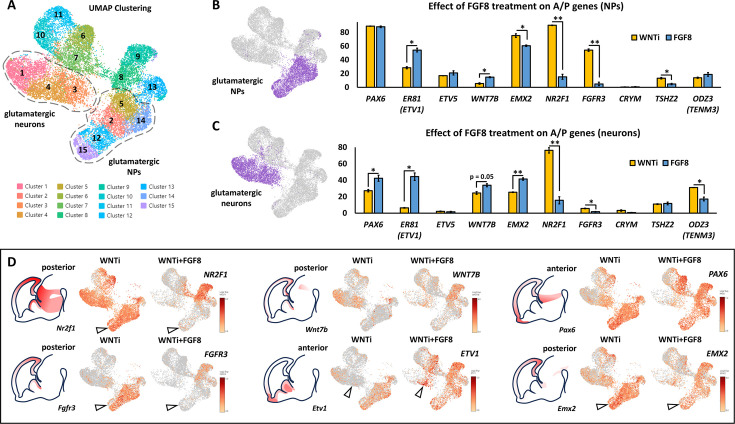
Figure 7. FGF8-dependent acquisition of distinct antero-posterior areal identities in human telencephalic organoids.
(A) Glutamatergic neural progenitor (NPs; clusters 2/5/12/14/15) and glutamatergic neurons (clusters 1/3/4) are highlighted on the UMAP projection of day69 organoid scRNAseq data. (B,C) Images on the right display the clusters selected for analysis, as described in A. The graphs show the percentages of cells expressing anterior-posterior (A/P) cortical markers within the two highlighted cluster groups: NPs (top graph) and neurons (bottom graph). The percentage of cells positive for anterior markers (PAX6, ER81, ETV5) and posterior markers (WNT7B, EMX2, NR2F1, FGFR3, CRYM, TSHZ2, ODZ3) is shown in yellow for control (WNTi) organoids and in blue for FGF8-treated (WNTi + FGF8) organoids. (D) Expression level of key posterior (NR2F1, FGFR3, WNT7B, EMX2) and anterior (ETV1, PAX6) genes in UMAP projections of WNTi or WNTi + FGF8 day69 organoid samples, as indicated. Black arrowheads in the NR2F1 and FGFR3 UMAP projections point to decreased expression in proliferating glutamatergic progenitors upon FGF8 treatment, while arrowheads in the ETV1 UMAP projection indicate increased expression in FGF8-treated glutamatergic neurons. Brain schematics with gene expression patterns are based on embryonic day 13.5 staining data from the Mouse Allen Brain Atlas.